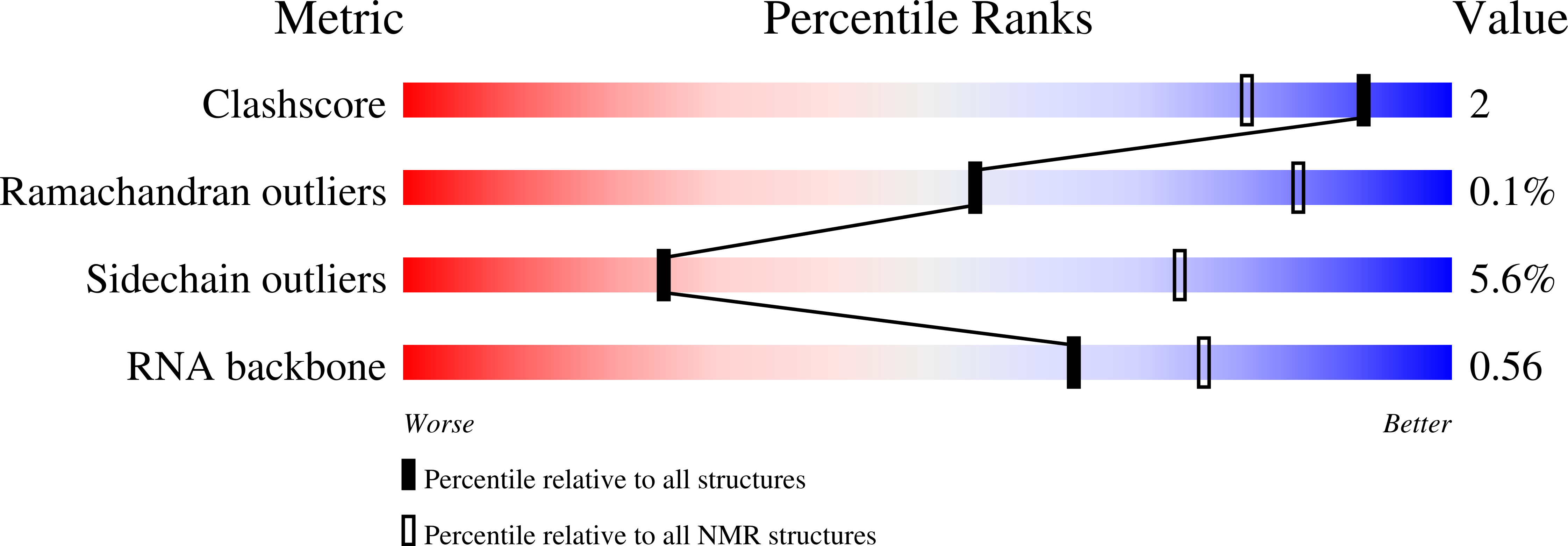
Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Duss, O., Michel, E., Diarra Dit Konte, N., Schubert, M., Allain, F.H.(2014) Nucleic Acids Res 42: 5332-5346
- PubMed: 24561806
- DOI: https://doi.org/10.1093/nar/gku141
- Primary Citation of Related Structures:
2MFC, 2MFE, 2MFF, 2MFG, 2MFH - PubMed Abstract:
The carbon storage regulator/regulator of secondary metabolism (Csr/Rsm) type of small non-coding RNAs (sRNAs) is widespread throughout bacteria and acts by sequestering the global translation repressor protein CsrA/RsmE from the ribosome binding site of a subset of mRNAs. Although we have previously described the molecular basis of a high affinity RNA target bound to RsmE, it remains unknown how other lower affinity targets are recognized by the same protein. Here, we have determined the nuclear magnetic resonance solution structures of five separate GGA binding motifs of the sRNA RsmZ of Pseudomonas fluorescens in complex with RsmE. The structures explain how the variation of sequence and structural context of the GGA binding motifs modulate the binding affinity for RsmE by five orders of magnitude (∼10 nM to ∼3 mM, Kd). Furthermore, we see that conformational adaptation of protein side-chains and RNA enable recognition of different RNA sequences by the same protein contributing to binding affinity without conferring specificity. Overall, our findings illustrate how the variability in the Csr/Rsm protein-RNA recognition allows a fine-tuning of the competition between mRNAs and sRNAs for the CsrA/RsmE protein.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland.