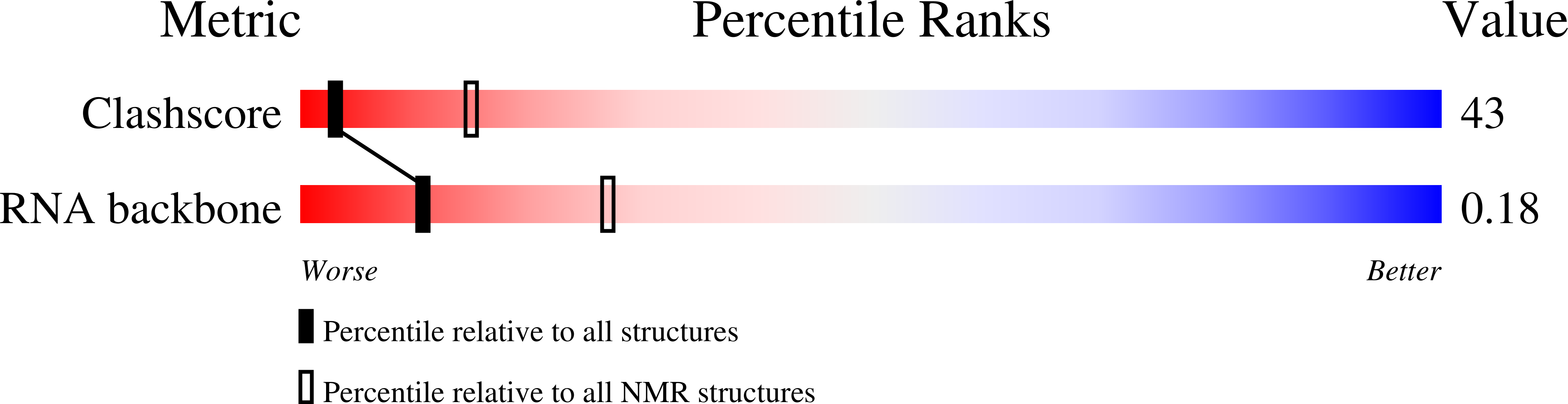Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function.
Richards, R.J., Wu, H., Trantirek, L., O'Connor, C.M., Collins, K., Feigon, J.(2006) RNA 12: 1475-1485
- PubMed: 16809815
- DOI: https://doi.org/10.1261/rna.112306
- Primary Citation of Related Structures:
2M21 - PubMed Abstract:
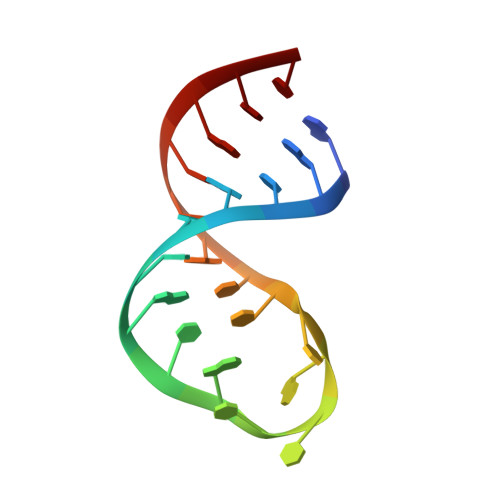
Tetrahymena telomerase RNA (TER) contains several regions in addition to the template that are important for function. Central among these is the stem-loop IV domain, which is involved in both catalysis and RNP assembly, and includes binding sites for both the holoenzyme assembly protein p65 and telomerase reverse transcriptase (TERT). Stem-loop IV contains two regions with high evolutionary sequence conservation: a central GA bulge between helices, and a terminal loop. We solved the solution structure of loop IV and modeled the structure of the helical region containing the GA bulge, using NMR and residual dipolar couplings. The central GA bulge with flanking C-G base pairs induces a approximately 50 degrees semi-rigid bend in the helix. Loop IV is highly structured, and contains a conserved C-U base pair at the top of the helical stem. Analysis of new and previous biochemical data in light of the structure provides a rationale for some of the sequence conservation in this region of TER. The results suggest that during holoenzyme assembly the protein p65 recognizes a bend in stem IV, and this binding to central stem IV helps to position the structured loop IV for interaction with TERT and other region(s) of TER.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of South Bohemia, Czech Republic.