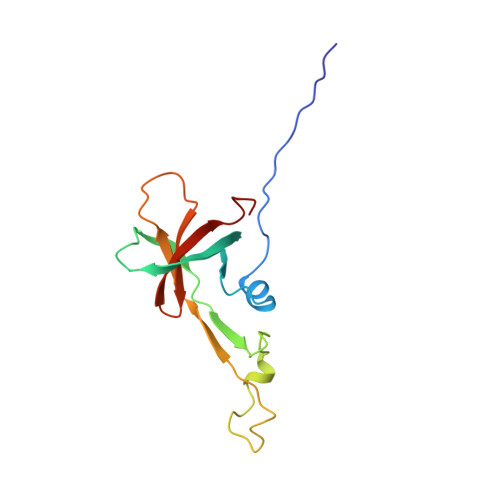
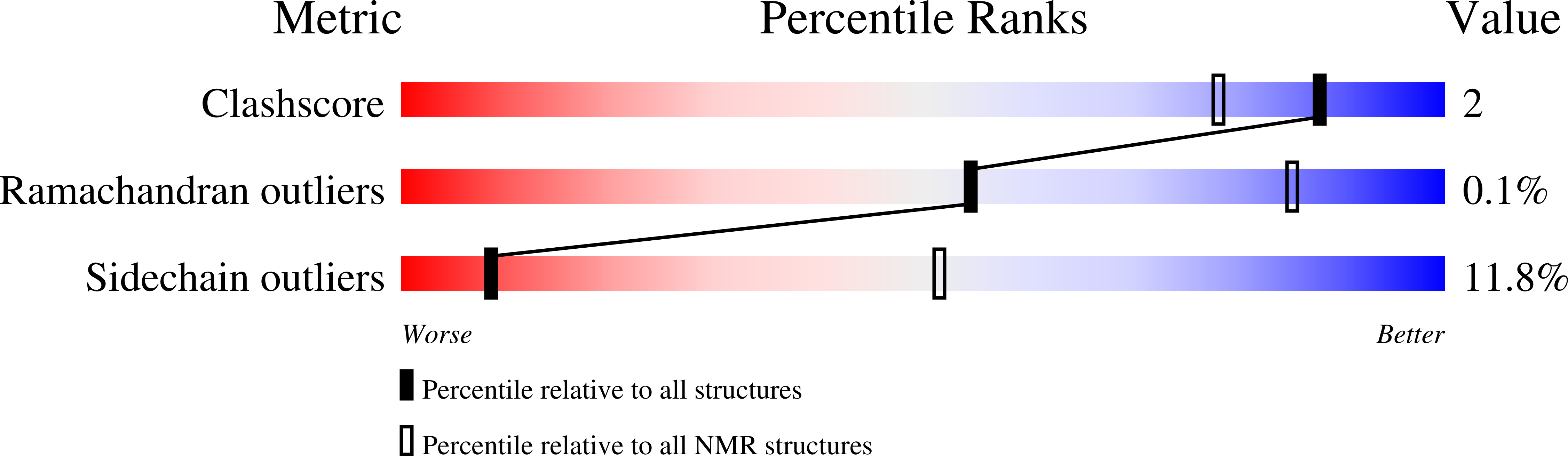The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
Christ, N.A., Bochmann, S., Gottstein, D., Duchardt-Ferner, E., Hellmich, U.A., Dusterhus, S., Kotter, P., Guntert, P., Entian, K.D., Wohnert, J.(2012) J Biol Chem 287: 35286-35298
- PubMed: 22904324
- DOI: https://doi.org/10.1074/jbc.M112.401620
- Primary Citation of Related Structures:
2LVL - PubMed Abstract:
Lantibiotics are peptide-derived antibiotics that inhibit the growth of Gram-positive bacteria via interactions with lipid II and lipid II-dependent pore formation in the bacterial membrane. Due to their general mode of action the Gram-positive producer strains need to express immunity proteins (LanI proteins) for protection against their own lantibiotics. Little is known about the immunity mechanism protecting the producer strain against its own lantibiotic on the molecular level. So far, no structures have been reported for any LanI protein. We solved the structure of SpaI, a LanI protein from the subtilin producing strain Bacillus subtilis ATCC 6633. SpaI is a 16.8-kDa lipoprotein that is attached to the outside of the cytoplasmic membrane via a covalent diacylglycerol anchor. SpaI together with the ABC transporter SpaFEG protects the B. subtilis membrane from subtilin insertion. The solution-NMR structure of a 15-kDa biologically active C-terminal fragment reveals a novel fold. We also demonstrate that the first 20 N-terminal amino acids not present in this C-terminal fragment are unstructured in solution and are required for interactions with lipid membranes. Additionally, growth tests reveal that these 20 N-terminal residues are important for the immunity mediated by SpaI but most likely are not part of a possible subtilin binding site. Our findings are the first step on the way of understanding the immunity mechanism of B. subtilis in particular and of other lantibiotic producing strains in general.
Organizational Affiliation:
Institute for Molecular Biosciences, Goethe University, 60438 Frankfurt am Main, Germany; Center of Biomolecular Magnetic Resonance, Goethe University, 60438 Frankfurt am Main, Germany.