NMR solution structure and biophysical characterization of Vibrio harveyi acyl carrier protein A75H: effects of divalent metal ions.
Chan, D.I., Chu, B.C., Lau, C.K., Hunter, H.N., Byers, D.M., Vogel, H.J.(2010) J Biol Chem 285: 30558-30566
- PubMed: 20659901
- DOI: https://doi.org/10.1074/jbc.M110.128298
- Primary Citation of Related Structures:
2L0Q - PubMed Abstract:
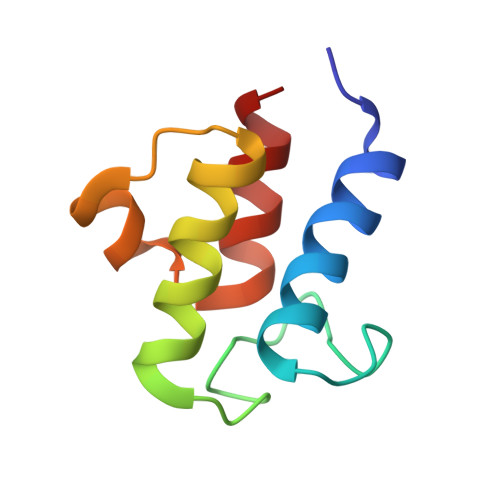
Bacterial acyl carrier protein (ACP) is a highly anionic, 9 kDa protein that functions as a cofactor protein in fatty acid biosynthesis. Escherichia coli ACP is folded at neutral pH and in the absence of divalent cations, while Vibrio harveyi ACP, which is very similar at 86% sequence identity, is unfolded under the same conditions. V. harveyi ACP adopts a folded conformation upon the addition of divalent cations such as Ca(2+) and Mg(2+) and a mutant, A75H, was previously identified that restores the folded conformation at pH 7 in the absence of divalent cations. In this study we sought to understand the unique folding behavior of V. harveyi ACP using NMR spectroscopy and biophysical methods. The NMR solution structure of V. harveyi ACP A75H displays the canonical ACP structure with four helices surrounding a hydrophobic core, with a narrow pocket closed off from the solvent to house the acyl chain. His-75, which is charged at neutral pH, participates in a stacking interaction with Tyr-71 in the far C-terminal end of helix IV. pH titrations and the electrostatic profile of ACP suggest that V. harveyi ACP is destabilized by anionic charge repulsion around helix II that can be partially neutralized by His-75 and is further reduced by divalent cation binding. This is supported by differential scanning calorimetry data which indicate that calcium binding further increases the melting temperature of V. harveyi ACP A75H by ∼20 °C. Divalent cation binding does not alter ACP dynamics on the ps-ns timescale as determined by (15)N NMR relaxation experiments, however, it clearly stabilizes the protein fold as observed by hydrogen-deuterium exchange studies. Finally, we demonstrate that the E. coli ACP H75A mutant is similarly unfolded as wild-type V. harveyi ACP, further stressing the importance of this particular residue for proper protein folding.
Organizational Affiliation:
Biochemistry Research Group, Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada.