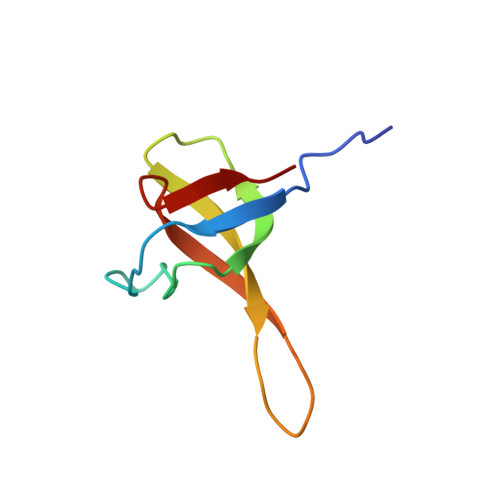
Chimeric SHA-D domain "SH3-Bergerac": 3D structure and dynamics studies
Khristoforov, V.S., Prokhorov, D.A., Timchenko, M.A., Kudrevatykh, Y.A., Gushchina, L.V., Filimonov, V.V., Kutyshenko, V.P.(2010) Russ J Bioorg Chem 36: 468-476
- PubMed: 20823919
- Primary Citation of Related Structures:
2KR3 - PubMed Abstract:
Protein SHA-D of "SH3-Bergerac" chimeric proteins family was constructed by substitution of beta-turn N47-D48 in spectrin SH3-domain by KATANDKTYE amino acid sequence. Structural and dynamics properties of SHA-D in solution were studied by with the help of high-resolution NMR. The extension of SHA-D polypeptide chain in comparison with wild type of protein WT-SH3 (~ 17%) practically doesn't affect almost the total molecule topology. 3D-structure of SHA-D is practically identical to the proteins of "SH3-Bergerac" family. However there are some differences in dynamic characteristics in the region of substitution. The G52D substitution in SHA-D protein results in a destabilization of the region insertion where the conditions for conformational exchange appear. Destabilization further affects the entire SHA- D molecule making its structure more labile.