Anti-Group A Streptococcal Vaccine Epitope: STRUCTURE, STABILITY, AND ITS ABILITY TO INTERACT WITH HLA CLASS II MOLECULES.
Guilherme, L., Alba, M.P., Ferreira, F.M., Oshiro, S.E., Higa, F., Patarroyo, M.E., Kalil, J.(2011) J Biol Chem 286: 6989-6998
- PubMed: 21169359
- DOI: https://doi.org/10.1074/jbc.M110.132118
- Primary Citation of Related Structures:
2KK9 - PubMed Abstract:
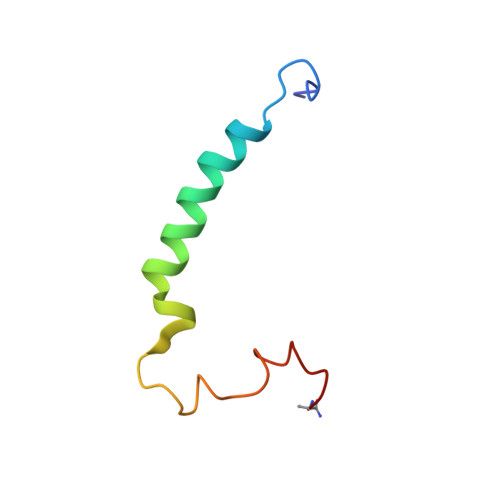
Streptococcus pyogenes infections remain a health problem in several countries due to poststreptococcal sequelae. We developed a vaccine epitope (StreptInCor) composed of 55 amino acids residues of the C-terminal portion of the M protein that encompasses both T and B cell protective epitopes. The nuclear magnetic resonance (NMR) structure of the StreptInCor peptide showed that the structure was composed of two microdomains linked by an 18-residue α-helix. A chemical stability study of the StreptInCor folding/unfolding process using far-UV circular dichroism showed that the structure was chemically stable with respect to pH and the concentration of urea. The T cell epitope is located in the first microdomain and encompasses 11 out of the 18 α-helix residues, whereas the B cell epitope is in the second microdomain and showed no α-helical structure. The prediction of StreptInCor epitope binding to different HLA class II molecules was evaluated based on an analysis of the 55 residues and the theoretical possibilities for the processed peptides to fit into the P1, P4, P6, and P9 pockets in the groove of several HLA class II molecules. We observed 7 potential sites along the amino acid sequence of StreptInCor that were capable of recognizing HLA class II molecules (DRB1*, DRB3*, DRB4*, and DRB5*). StreptInCor-overlapping peptides induced cellular and humoral immune responses of individuals bearing different HLA class II molecules and could be considered as a universal vaccine epitope.
Organizational Affiliation:
Heart Institute (InCor), University of São Paulo, São Paulo 5403-903, Brazil. luizagui@usp.br