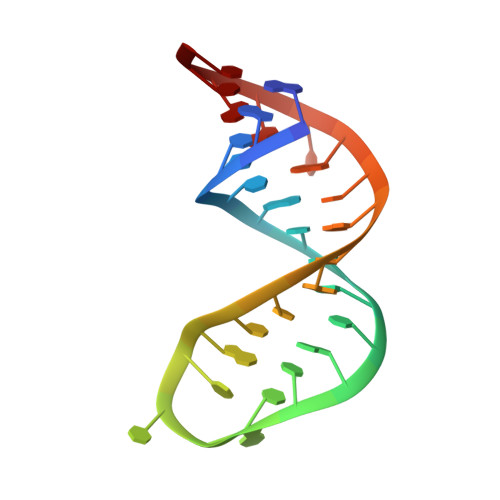
The unstable part of the apical stem of duck hepatitis B virus epsilon shows enhanced base pair opening but not pico- to nanosecond dynamics and is essential for reverse transcriptase binding.
Ampt, K.A., van der Werf, R.M., Nelissen, F.H., Tessari, M., Wijmenga, S.S.(2009) Biochemistry 48: 10499-10508
- PubMed: 19817488
- DOI: https://doi.org/10.1021/bi9011385
- Primary Citation of Related Structures:
2K5Z - PubMed Abstract:
Hepatitis B virus (HBV) replication starts with binding of reverse transcriptase (RT) to the apical stem-loop region of epsilon, a conserved element of the RNA pregenome. For duck HBV, an in vitro replication system has provided molecular details of this interaction. Further insights can be obtained from the structure and dynamics of the duck and human apical stem-loops. Previously, we reported these for the human apical stem-loop. Here, we present the same for the duck counterpart. Unlike its human counterpart, the duck apical stem is unstable in its middle/upper part and contains noncanonical base pairs. This dynamics study is the first of an unstable RNA-DNA stem. Similar to the human stem, the duck apical stem comprises two helical segments with a bend angle of ca. 10 degrees , separated by a nonpaired mobile U residue. It is capped by a well-structured conserved UGUU loop with two residues mobile on the pico- to nanosecond time scale, one of which is involved in RT binding. Remarkably, the unstable middle/upper part of the stem does not show enhanced pico- to nanosecond time scale dynamics. Instead, adenine dispersion relaxation studies indicate enhanced millisecond time scale dynamics involving base pair opening. It can then be concluded that base pair opening is essential for epsilon-RT binding, because stabilization of the stem abolishes binding. We hypothesize that binding occurs by conformational capture of bases in the base pair open state. The unstable secondary structure of the apical stem-loop makes duck epsilon-RT binding unusual in light of recent classifications of RNA target interactions that assume stable secondary structures.
Organizational Affiliation:
Biophysical Chemistry, Institute of Molecules and Materials, Radboud University of Nijmegen, Heyendaalseweg 135, 6525 AJ Nijmegen, The Netherlands.