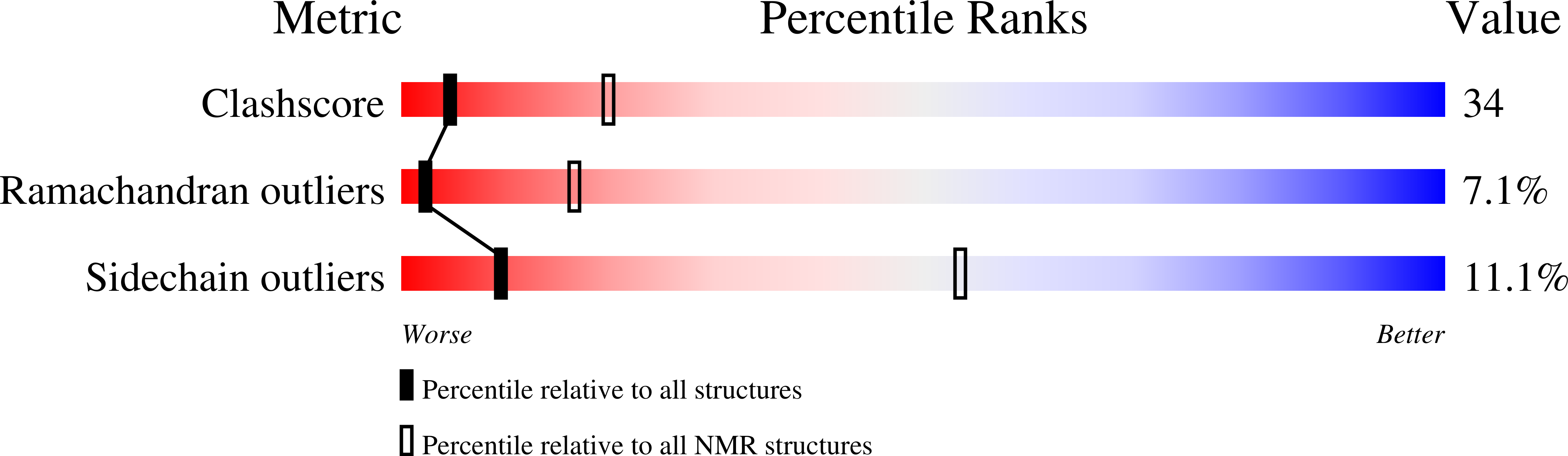NMR structure of a phosphatidyl-ethanolamine binding protein from Drosophila
Rautureau, G.J.P., Vovelle, F., Schoentgen, F., Decoville, M., Locker, D., Damblon, C., Jouvensal, L.(2009) Proteins 78: 1606-1610
- PubMed: 20131378
- DOI: https://doi.org/10.1002/prot.22682
- Primary Citation of Related Structures:
2JYZ
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville Victoria, Australia.