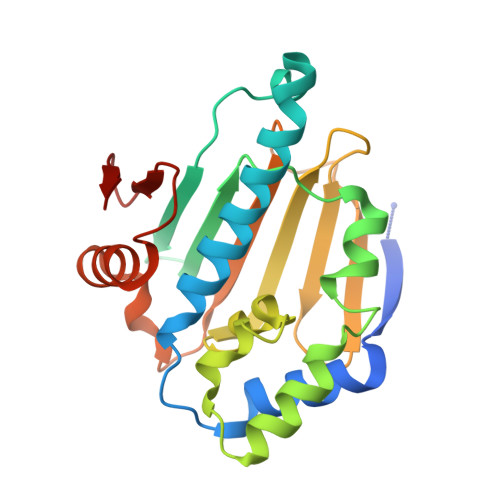
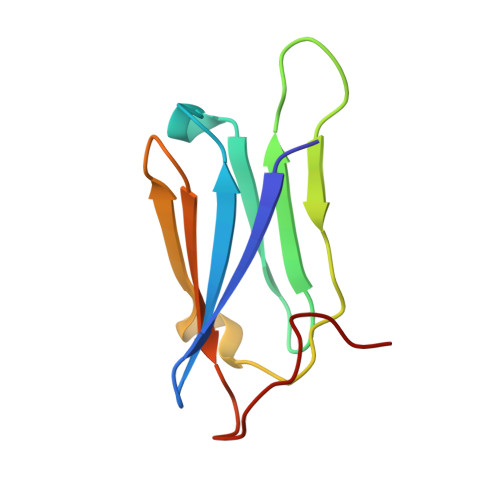
Structural and Functional Coupling of Hsp90- and Sgt1-Centred Multi-Protein Complexes.
Zhang, M., Boter, M., Li, K., Kadota, Y., Panaretou, B., Prodromou, C., Shirasu, K., Pearl, L.H.(2008) EMBO J 27: 2789
- PubMed: 18818696
- DOI: https://doi.org/10.1038/emboj.2008.190
- Primary Citation of Related Structures:
2JKI - PubMed Abstract:
Sgt1 is an adaptor protein implicated in a variety of processes, including formation of the kinetochore complex in yeast, and regulation of innate immunity systems in plants and animals. Sgt1 has been found to associate with SCF E3 ubiquitin ligases, the CBF3 kinetochore complex, plant R proteins and related animal Nod-like receptors, and with the Hsp90 molecular chaperone. We have determined the crystal structure of the core Hsp90-Sgt1 complex, revealing a distinct site of interaction on the Hsp90 N-terminal domain. Using the structure, we developed mutations in Sgt1 interfacial residues, which specifically abrogate interaction with Hsp90, and disrupt Sgt1-dependent functions in vivo, in plants and yeast. We show that Sgt1 bridges the Hsp90 molecular chaperone system to the substrate-specific arm of SCF ubiquitin ligase complexes, suggesting a role in SCF assembly and regulation, and providing multiple complementary routes for ubiquitination of Hsp90 client proteins.
Organizational Affiliation:
Section of Structural Biology, The Institute of Cancer Research, Chester Beatty Laboratories, London, UK.