Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein
Crabtree, B., Holloway, D.E., Baker, M.D., Acharya, K.R., Subramanian, V.(2007) Biochemistry 46: 2431
- PubMed: 17279775
- DOI: https://doi.org/10.1021/bi062158n
- Primary Citation of Related Structures:
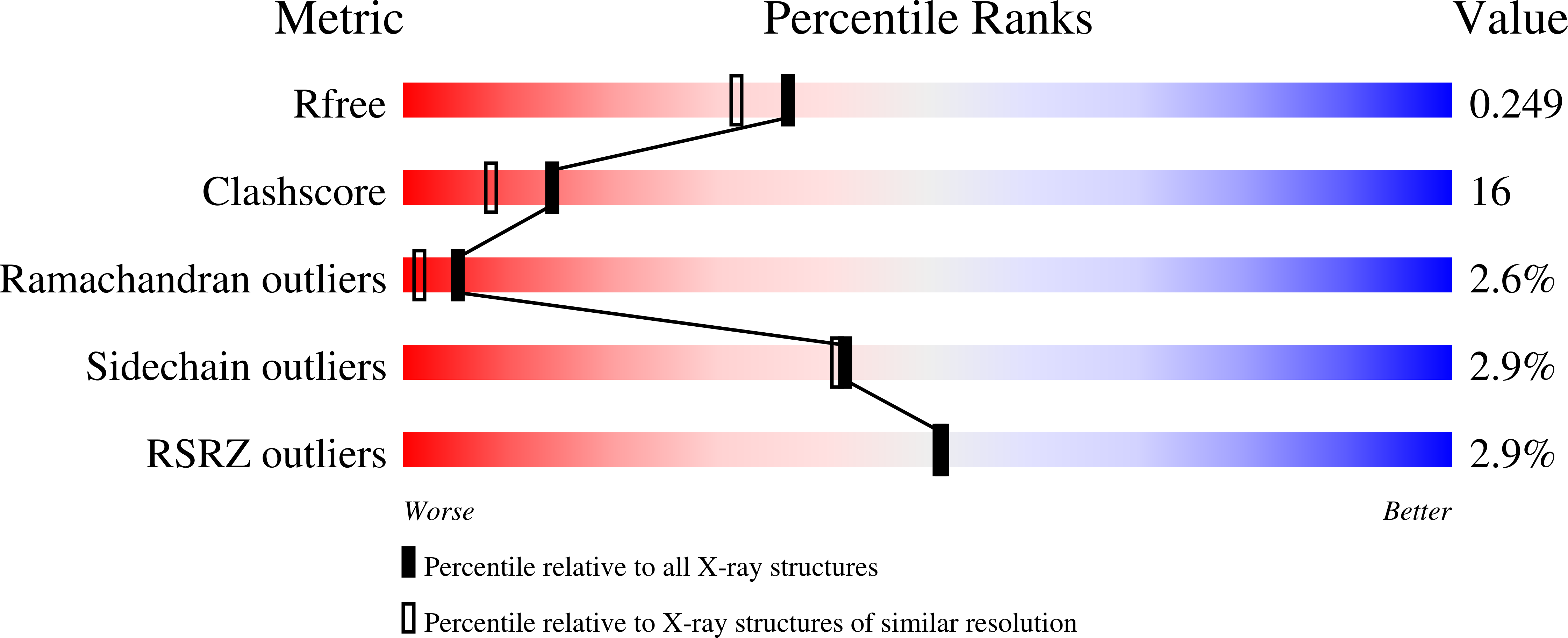
2J4T - PubMed Abstract:
Murine angiogenin-4 (mAng-4) is a member of the pancreatic ribonuclease superfamily that is expressed in some endodermally derived organs. We now show that mAng-4 is angiogenic using a thoracic aorta assay never before applied to the angiogenins. mAng-4, human angiogenin (hAng), and murine angiogenin-1 (mAng-1) stimulate the proliferation of IGR1 melanoma cells but do not stimulate the proliferation or migration of bovine corneal endothelial cells or primary mouse embryonic fibroblasts. In addition, we report the 3-D structure of mAng-4 at 2.02-A resolution. The structure shows that the residues forming the putative B1, P1, and B2 RNA-binding subsites occupy positions similar to their hAng counterparts. The B1 subsite is obstructed by Glu115 and Ile118. The obstruction is stabilized by a novel salt bridge between the C-terminal carboxyl group and the side chain of Arg99. Through mutational studies, we identify residues critical to the angiogenic function of mAng-4. The effect of H12A and H112A mutations in the catalytic site indicates that ribonucleolytic activity is essential to angiogenesis. The consequences of a nearby E115A mutation are consistent with a significant role for Glu115 in the attenuation of enzymatic activity but also suggest that sufficient suppression of catalysis is necessary for angiogenesis. The effect of an R32A mutation in the putative nuclear localization sequence indicates that this residue is crucial for angiogenesis. In the putative cell-binding segment, the replacement of Lys59 with Asn (its counterpart at position 61 of hAng) does not abrogate enzymatic activity but abolishes angiogenic activity, the reason for which is unclear.
Organizational Affiliation:
Department of Biology and Biochemistry, University of Bath, Claverton Down, Bath BA2 7AY, United Kingdom.