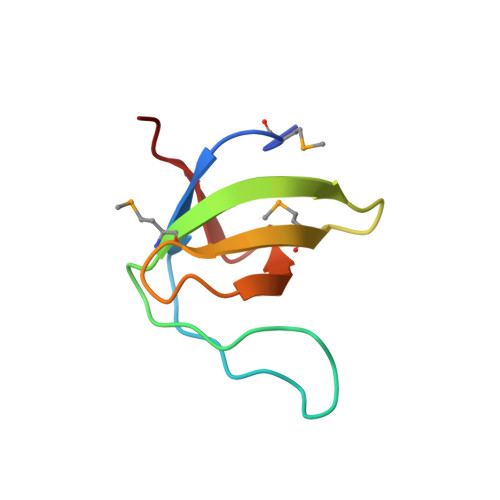
High Resolution Crystal Structures of the P120 Rasgap SH3 Domain.
Ross, B., Kristensen, O., Favre, D., Walicki, J., Kastrup, J.S., Widmann, C., Gajhede, M.(2007) Biochem Biophys Res Commun 353: 463
- PubMed: 17188236
- DOI: https://doi.org/10.1016/j.bbrc.2006.12.044
- Primary Citation of Related Structures:
2J05, 2J06 - PubMed Abstract:
X-ray structures of two crystal forms of the Src homology 3 domain (SH3) of the Ras GTPase activating protein (RasGAP) were determined at 1.5 and 1.8A resolution. The overall structure comprises a single domain with two tightly packed beta-sheets linked by a short helical segment. An important motif for peptide binding in other SH3 domains is not conserved in RasGAP. The RasGAP SH3 domain forms dimers in the crystal structures, which may provide new functional insight. The dimer interface involves residues also present in a peptide previously identified as an apoptotic sensitizer of tumor cells.
Organizational Affiliation:
Biostructural Research, Department of Medicinal Chemistry, The Danish University of Pharmaceutical Sciences, Universitetsparken 2, DK-2100 Copenhagen, Denmark.