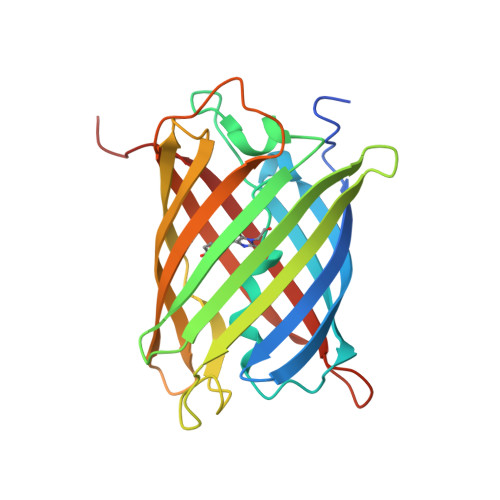
Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Pletneva, N., Pletnev, V., Tikhonova, T., Pakhomov, A.A., Popov, V., Martynov, V.I., Wlodawer, A., Dauter, Z., Pletnev, S.(2007) Acta Crystallogr D Biol Crystallogr 63: 1082-1093
- PubMed: 17881826
- DOI: https://doi.org/10.1107/S0907444907042461
- Primary Citation of Related Structures:
2ICR, 2OJK, 2PXS, 2PXW - PubMed Abstract:
The three-dimensional structures of the wild-type red (zRFP574) and green (zGFP506) fluorescent proteins (FP) from the button polyp Zoanthus have been determined at 1.51 and 2.2 A resolution, respectively. In addition, the crystal structures of a zGFP506 variant (zGFP506_N66D) with replacement of the first chromophore-forming residue (Asn66 to Asp) have been determined in the transitional 'green' and mature 'red' states at 2.4 and 2.2 A, respectively. The monomers of these proteins adopt the typical fold of the green fluorescent protein (GFP) family, consisting of an 11-stranded beta-barrel with a chromophore embedded in the middle of an internal alpha-helix directed along the beta-barrel axis. Post-translational modification of the chromophore-forming sequence Asn66-Tyr67-Gly68 within zGFP506 results in a typical GFP-like coplanar two-ring structure consisting of a five-membered imidazolinone heterocycle with the phenolic ring of Tyr67 in a cis orientation to the C(alpha)-N(67) bond. A novel post-translational modification of the chromophore-forming sequence Asp66-Tyr67-Gly68 in zRFP574 expands the protein maturation beyond the green-emitting form and results in decarboxylation of the Asp66 side chain. It is suggested that electrostatic conflict between the closely spaced negatively charged side chains of the chromophore Asp66 and the proximal catalytic Glu221 is most likely to be the trigger for the chain of reactions resulting in the observed decarboxylation. The chromophore structures of wild-type zGFP506 and of its mutant zGFP506_N66D in the 'green' and 'red' states support this suggestion. The beta-barrel frames of zRFP574 and zGFP506 reveal the presence of a water-filled pore leading to the chromophore Tyr67, similar to that observed previously in TurboGFP. An analysis of the residue composition at two inter-monomer interfaces in the tetrameric biological unit of zRFP574 and zGFP506, as well as of zYFP538 from the same species, has revealed a group of highly conserved residues that are apparently responsible for oligomerization. These residues present initial useful targets for rational mutagenesis aimed at designing monomeric forms of the fluorescent proteins, which are more suitable for practical applications.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Science, Moscow, Russia.