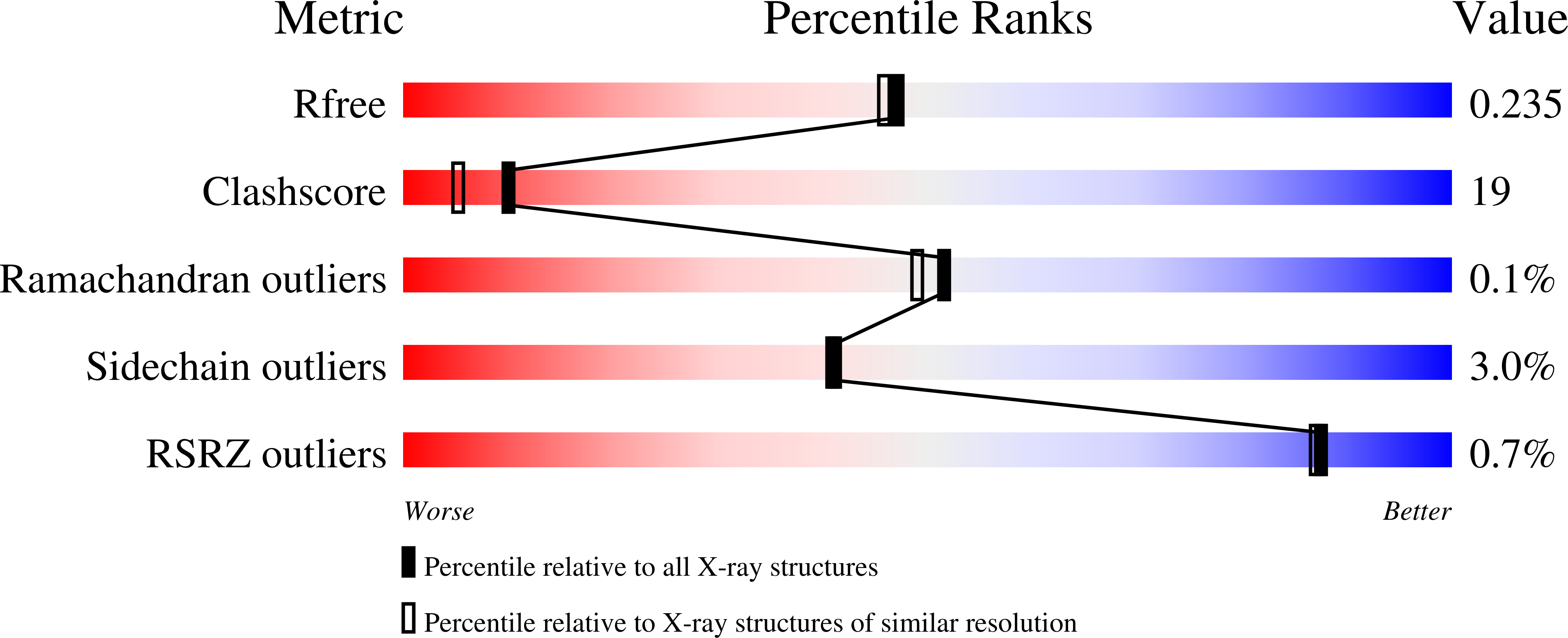
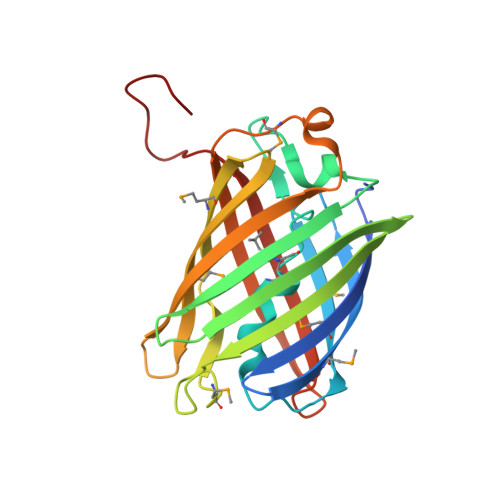
Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea Anemone Cnidopus Japonicus
Chan, M.C.Y., Karasawa, S., Mizuno, H., Bosanac, I., Ho, D., Prive, G.G., Miyawaki, A., Ikura, M.(2006) J Biol Chem 281: 37813-37819
- PubMed: 17028187
- DOI: https://doi.org/10.1074/jbc.M606921200
- Primary Citation of Related Structures:
2IB5, 2IB6 - PubMed Abstract:
Green fluorescent protein (GFP) and its relatives (GFP protein family) have been isolated from marine organisms such as jellyfish and corals that belong to the phylum Cnidaria (stinging aquatic invertebrates). They are intrinsically fluorescent proteins. In search of new members of the family of green fluorescent protein family, we identified a non-fluorescent chromoprotein from the Cnidopus japonicus species of sea anemone that possesses 45% sequence identity to dsRed (a red fluorescent protein). This newly identified blue color protein has an absorbance maximum of 610 nm and is hereafter referred to as cjBlue. Determination of the cjBlue 1.8 A crystal structure revealed a chromophore comprised of Gln(63)-Tyr(64)-Gly(65). The ring stacking between Tyr(64) and His(197) stabilized the cjBlue trans chromophore conformation along the Calpha2-Cbeta2 bond of 5-[(4-hydroxyphenyl)methylene]-imidazolinone, which closely resembled that of the "Kindling Fluorescent Protein" and Rtms5. Replacement of Tyr(64) with Leu in wild-type cjBlue produced a visible color change from blue to yellow with a new absorbance maximum of 417 nm. Interestingly, the crystal structure of the yellow mutant Y64L revealed two His(197) imidazole ring orientations, suggesting a flip-flop interconversion between the two conformations in solution. We conclude that the dynamics and structure of the chromophore are both essential for the optical appearance of these color proteins.
Organizational Affiliation:
Division of Signaling Biology, Ontario Cancer Institute and Department of Medical Biophysics, University of Toronto, Toronto, Ontario M5G 1L7, Canada.