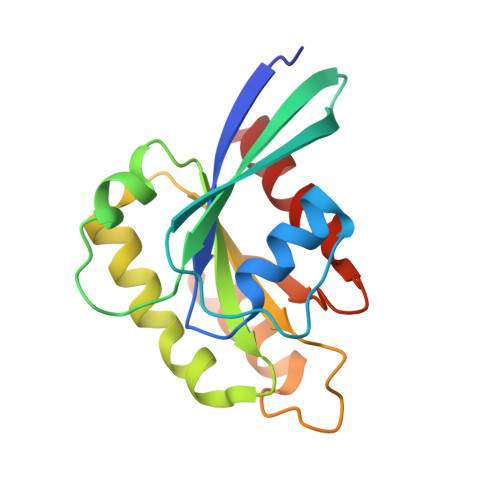
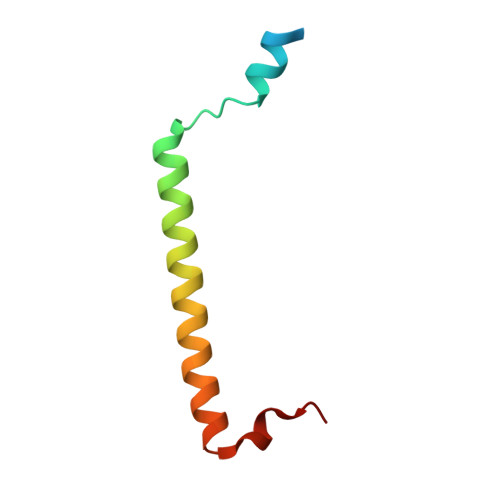
Structural Basis for Rab11-mediated Recruitment of FIP3 to Recycling Endosomes.
Eathiraj, S., Mishra, A., Prekeris, R., Lambright, D.G.(2006) J Mol Biol 364: 121-135
- PubMed: 17007872
- DOI: https://doi.org/10.1016/j.jmb.2006.08.064
- Primary Citation of Related Structures:
2HV8 - PubMed Abstract:
The Rab11 GTPase regulates recycling of internalized plasma membrane receptors and is essential for completion of cytokinesis. A family of Rab11 interacting proteins (FIPs) that conserve a C-terminal Rab-binding domain (RBD) selectively recognize the active form of Rab11. Normal completion of cytokinesis requires a complex between Rab11 and FIP3. Here, we report the crystal structure and mutational analysis of a heterotetrameric complex between constitutively active Rab11 and a FIP3 construct that includes the RBD. Two Rab11 molecules bind to dyad symmetric sites at the C terminus of FIP3, which forms a non-canonical coiled-coiled dimer with a flared C terminus and hook region. The RBD overlaps with the coiled coil and extends through the C-terminal hook. Although FIP3 engages the switch and interswitch regions of Rab11, the mode of interaction differs significantly from that of other Rab-effector complexes. In particular, the switch II region undergoes a large structural rearrangement from an ordered but non-complementary active conformation to a remodeled conformation that facilitates the interaction with FIP3. Finally, we provide evidence that FIP3 can form homo-oligomers in cells, and that a critical determinant of Rab11 binding in vitro is necessary for FIP3 recruitment to recycling endosomes during cytokinesis.
Organizational Affiliation:
Program in Molecular Medicine and Department of Biochemistry & Molecular Pharmacology, University of Massachusetts Medical School, Worcester, MA 01605, USA.