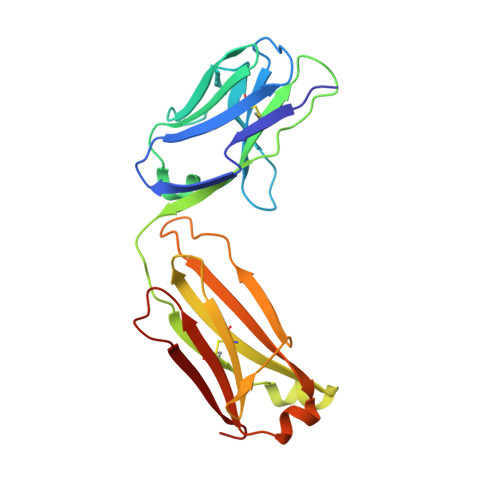
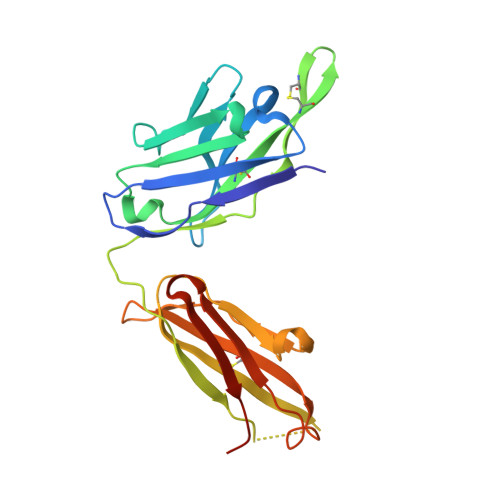
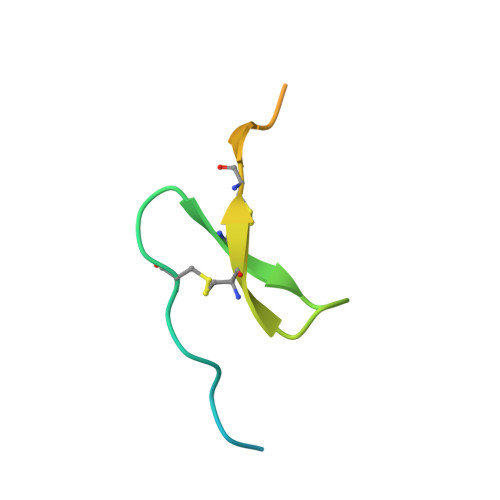
Synthetic anti-BR3 antibodies that mimic BAFF binding and target both human and murine B cells.
Lee, C.V., Hymowitz, S.G., Wallweber, H.J., Gordon, N.C., Billeci, K.L., Tsai, S.P., Compaan, D.M., Yin, J., Gong, Q., Kelley, R.F., Deforge, L.E., Martin, F., Starovasnik, M.A., Fuh, G.(2006) Blood 108: 3103-3111
- PubMed: 16840730
- DOI: https://doi.org/10.1182/blood-2006-03-011031
- Primary Citation of Related Structures:
2HFF, 2HFG - PubMed Abstract:
BR3, which is expressed on all mature B cells, is a specific receptor for the B-cell survival and maturation factor BAFF (B-cell-activating factor belonging to the tumor necrosis factor [TNF] family). In order to investigate the consequences of targeting BR3 in murine models and to assess the potential of BR3 antibodies as human therapeutics, synthetic antibody phage libraries were employed to identify BAFF-blocking antibodies cross-reactive to murine and human BR3, which share 52% identity in their extracellular domains. We found an antibody, CB1, which exhibits muM affinity for murine BR3 and very weak affinity for the human receptor. CB3s, an affinity-matured variant of CB1, has sub-nM affinity for BR3 from both species. Alanine scanning and crystallographic structural analysis of the CB3s/BR3 complex reveal that CB3s mimics BAFF by interacting with a similar region of the BR3 surface. Despite this similarity in binding epitopes, CB1 variants antagonize BAFF-dependent human B-cell proliferation in vitro and are effective at reducing murine B-cell populations in vivo, showing significant promise as therapeutics for human B-cell-mediated diseases.
Organizational Affiliation:
Department of Protein Engineering, Genentech Inc, 1 DNA Way, South San Francisco, CA 94080, USA.