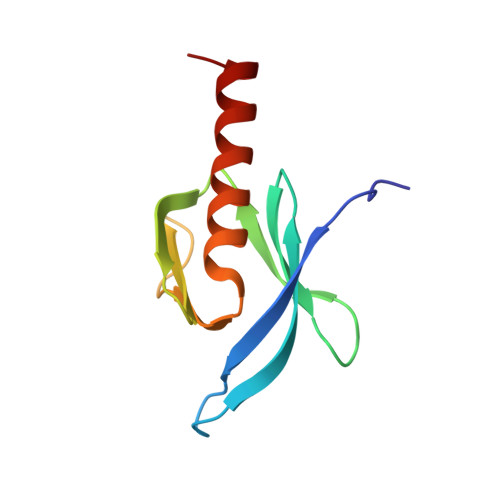
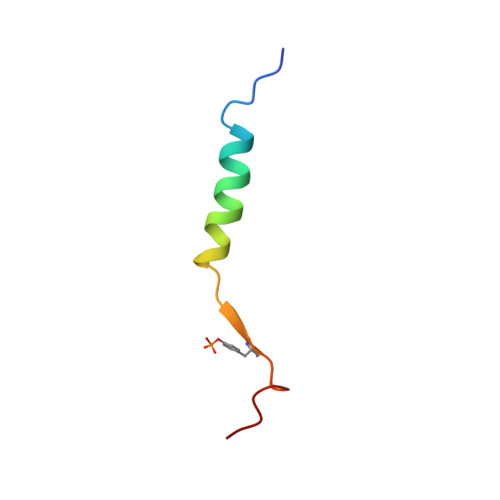
Structural basis of integrin activation by talin
Wegener, K.L., Partridge, A.W., Han, J., Pickford, A.R., Liddington, R.C., Ginsberg, M.H., Campbell, I.D.(2007) Cell 128: 171-182
- PubMed: 17218263
- DOI: https://doi.org/10.1016/j.cell.2006.10.048
- Primary Citation of Related Structures:
2H7D, 2H7E - PubMed Abstract:
Regulation of integrin affinity (activation) is essential for metazoan development and for many pathological processes. Binding of the talin phosphotyrosine-binding (PTB) domain to integrin beta subunit cytoplasmic domains (tails) causes activation, whereas numerous other PTB-domain-containing proteins bind integrins without activating them. Here we define the structure of a complex between talin and the membrane-proximal integrin beta3 cytoplasmic domain and identify specific contacts between talin and the integrin tail required for activation. We used structure-based mutagenesis to engineer talin and beta3 variants that interact with comparable affinity to the wild-type proteins but inhibit integrin activation by competing with endogenous talin. These results reveal the structural basis of talin's unique ability to activate integrins, identify an interaction that could aid in the design of therapeutics to block integrin activation, and enable engineering of cells with defects in the activation of multiple classes of integrins.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, England, UK.