Structural Insight into Pre-B Cell Receptor Function
Bankovich, A.J., Raunser, S., Juo, Z.S., Walz, T., Davis, M.M., Garcia, K.C.(2007) Science 316: 291-294
- PubMed: 17431183
- DOI: https://doi.org/10.1126/science.1139412
- Primary Citation of Related Structures:
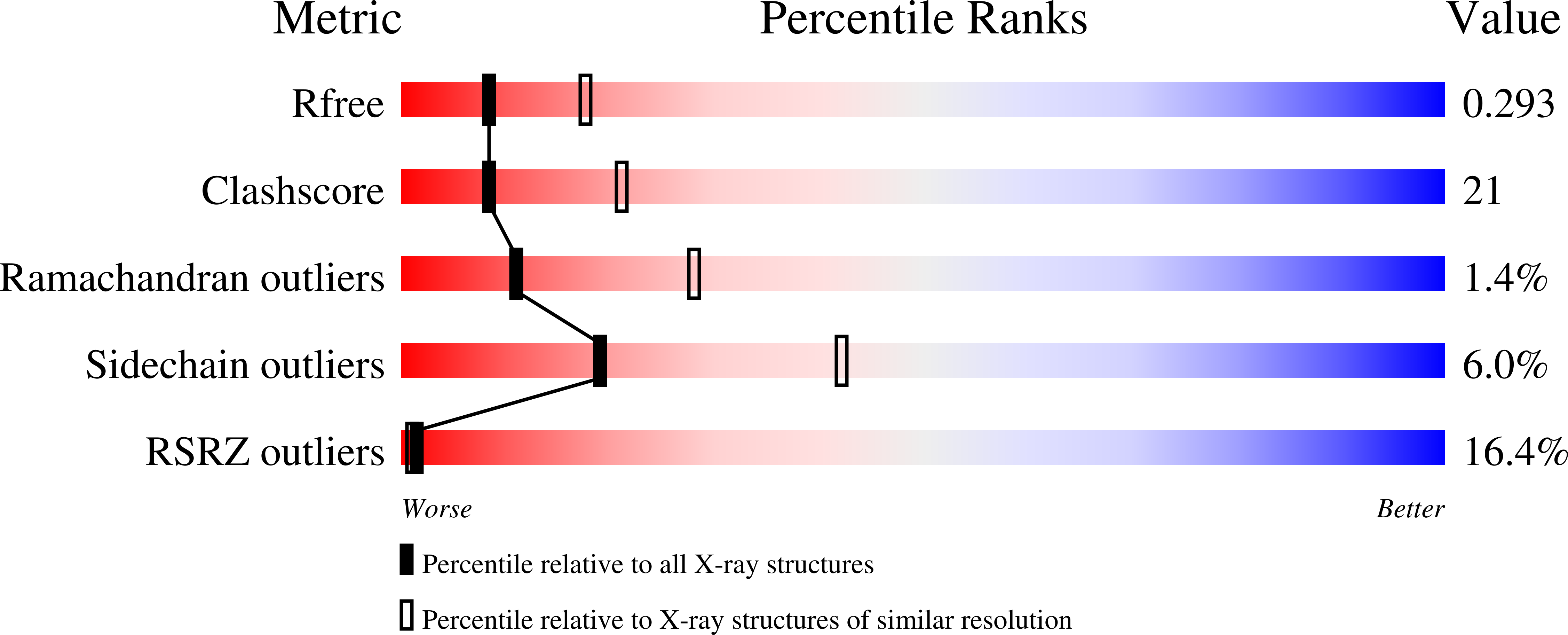
2H32, 2H3N - PubMed Abstract:
The pre-B cell receptor (pre-BCR) serves as a checkpoint in B cell development. In the 2.7 angstrom structure of a human pre-BCR Fab-like fragment, consisting of an antibody heavy chain (HC) paired with the surrogate light chain, the "unique regions" of VpreB and lambda5 replace the complementarity-determining region 3 (CDR3) loop of an antibody light chain and appear to "probe" the HC CDR3, potentially influencing the selection of the antibody repertoire. Biochemical analysis indicates that the pre-BCR is impaired in its ability to recognize antigen, which, together with electron microscopic visualization of a pre-BCR dimer, suggests ligand-independent oligomerization as the likely signaling mechanism.
Organizational Affiliation:
Program in Immunology, Stanford University School of Medicine, Stanford, CA 94305, USA.