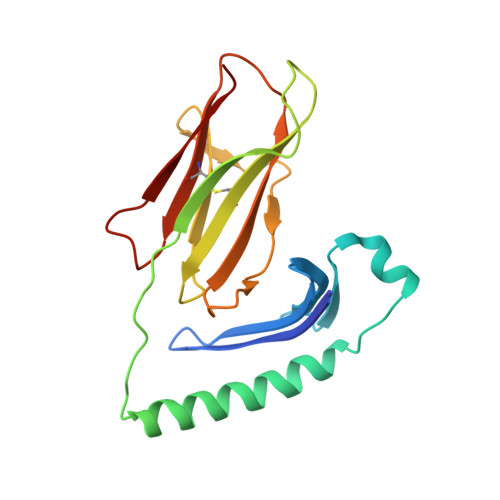
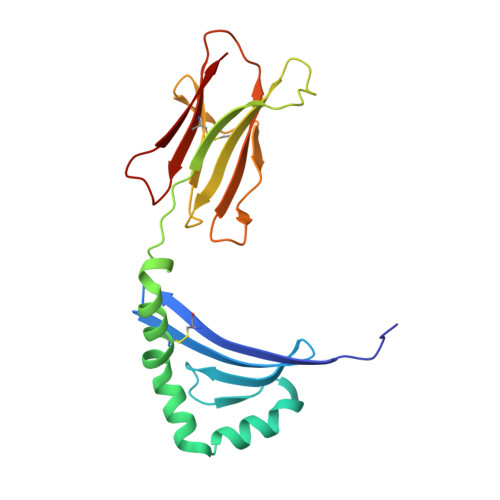
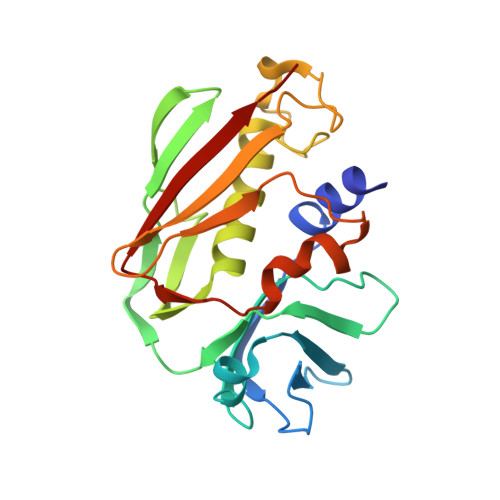
Crystal structure of staphylococcal enterotoxin I (SEI) in complex with a human major histocompatibility complex class II molecule.
Fernandez, M.M., Guan, R., Swaminathan, C.P., Malchiodi, E.L., Mariuzza, R.A.(2006) J Biol Chem 281: 25356-25364
- PubMed: 16829512
- DOI: https://doi.org/10.1074/jbc.M603969200
- Primary Citation of Related Structures:
2G9H - PubMed Abstract:
Superantigens are bacterial or viral proteins that elicit massive T cell activation through simultaneous binding to major histocompatibility complex (MHC) class II and T cell receptors. This activation results in uncontrolled release of inflammatory cytokines, causing toxic shock. A remarkable property of superantigens, which distinguishes them from T cell receptors, is their ability to interact with multiple MHC class II alleles independently of MHC-bound peptide. Previous crystallographic studies have shown that staphylococcal and streptococcal superantigens belonging to the zinc family bind to a high affinity site on the class II beta-chain. However, the basis for promiscuous MHC recognition by zinc-dependent superantigens is not obvious, because the beta-chain is polymorphic and the MHC-bound peptide forms part of the binding interface. To understand how zinc-dependent superantigens recognize MHC, we determined the crystal structure, at 2.0 A resolution, of staphylococcal enterotoxin I bound to the human class II molecule HLA-DR1 bearing a peptide from influenza hemagglutinin. Interactions between the superantigen and DR1 beta-chain are mediated by a zinc ion, and 22% of the buried surface of peptide.MHC is contributed by the peptide. Comparison of the staphylococcal enterotoxin I.peptide.DR1 structure with ones determined previously revealed that zinc-dependent superantigens achieve promiscuous binding to MHC by targeting conservatively substituted residues of the polymorphic beta-chain. Additionally, these superantigens circumvent peptide specificity by engaging MHC-bound peptides at their conformationally conserved N-terminal regions while minimizing sequence-specific interactions with peptide residues to enhance cross-reactivity.
Organizational Affiliation:
Instituto de Estudios de la Inmunidad Humoral, Laboratorio de Inmunología Estructural, Consejo Nacional de Investigaciones Científicas y Técnicas, Facultad de Farmacia y Bioquímica, Universidad de Buenos Aires, Buenos Aires, Argentina.