Structure of a red fluorescent protein from Zoanthus, zRFP574, reveals a novel chromophore
Pletneva, N., Pletnev, S., Tikhonova, T., Popov, V., Martynov, V., Pletnev, V.(2006) Acta Crystallogr D Biol Crystallogr 62: 527-532
- PubMed: 16627946
- DOI: https://doi.org/10.1107/S0907444906007852
- Primary Citation of Related Structures:
2FL1 - PubMed Abstract:
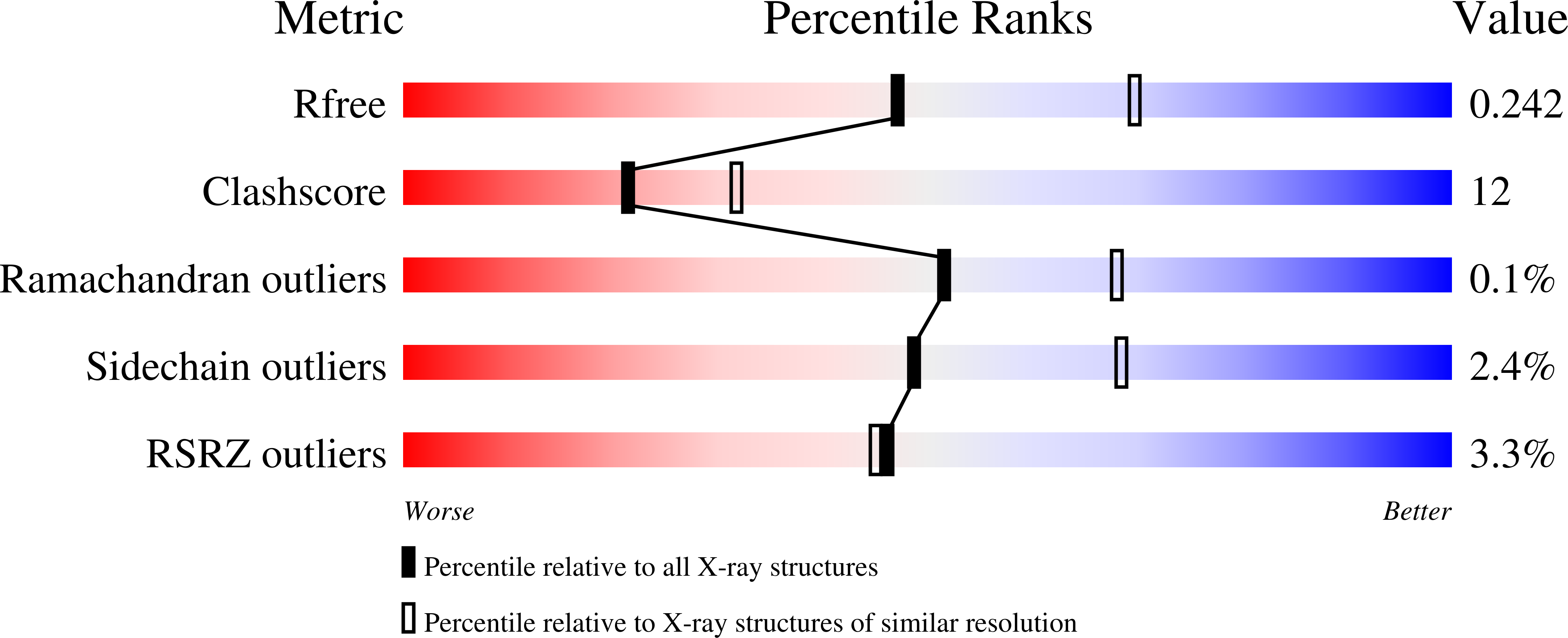
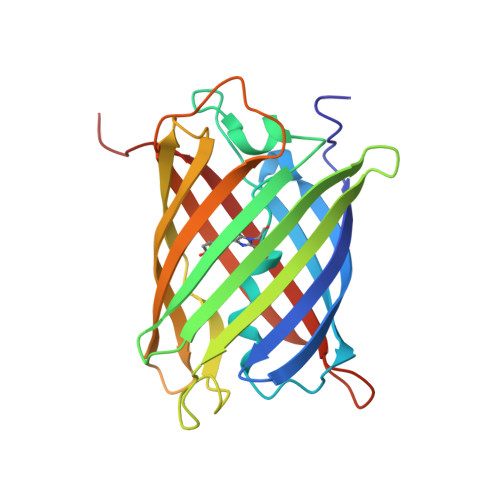
The three-dimensional structure of the red fluorescent protein (RFP) zRFP574 from the button polyp Zoanthus sp. (two dimers per asymmetric unit, 231 x 4 amino acids) has been determined at 2.4 A resolution in space group C222(1). The crystal structure, refined to a crystallographic R factor of 0.203 (R(free) = 0.249), adopts the beta-barrel fold composed of 11 strands similar to that of the yellow fluorescent protein zYFP538. The zRFP574 chromophore, originating from the protein sequence Asp66-Tyr67-Gly68, has a two-ring structure typical of GFP-like proteins. The bond geometry of residue 66 shows the strong tendency of the corresponding C(alpha) atom to sp(2) hybridization as a consequence of N-acylimine bond formation. The zRFP574 chromophore contains the 65-66 cis-peptide bond characteristic of red fluorescent proteins. The chromophore phenolic ring adopts a cis conformation coplanar with the imidazolinone ring. The crystallographic study has revealed an unexpected chemical feature of the internal chromophore. A decarboxylated side chain of the chromophore-forming residue Asp66 has been observed in the structure. This additional post-translational modification is likely to play a key role in the bathochromic shift of the zRFP574 spectrum.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Science, Moscow, Russia.