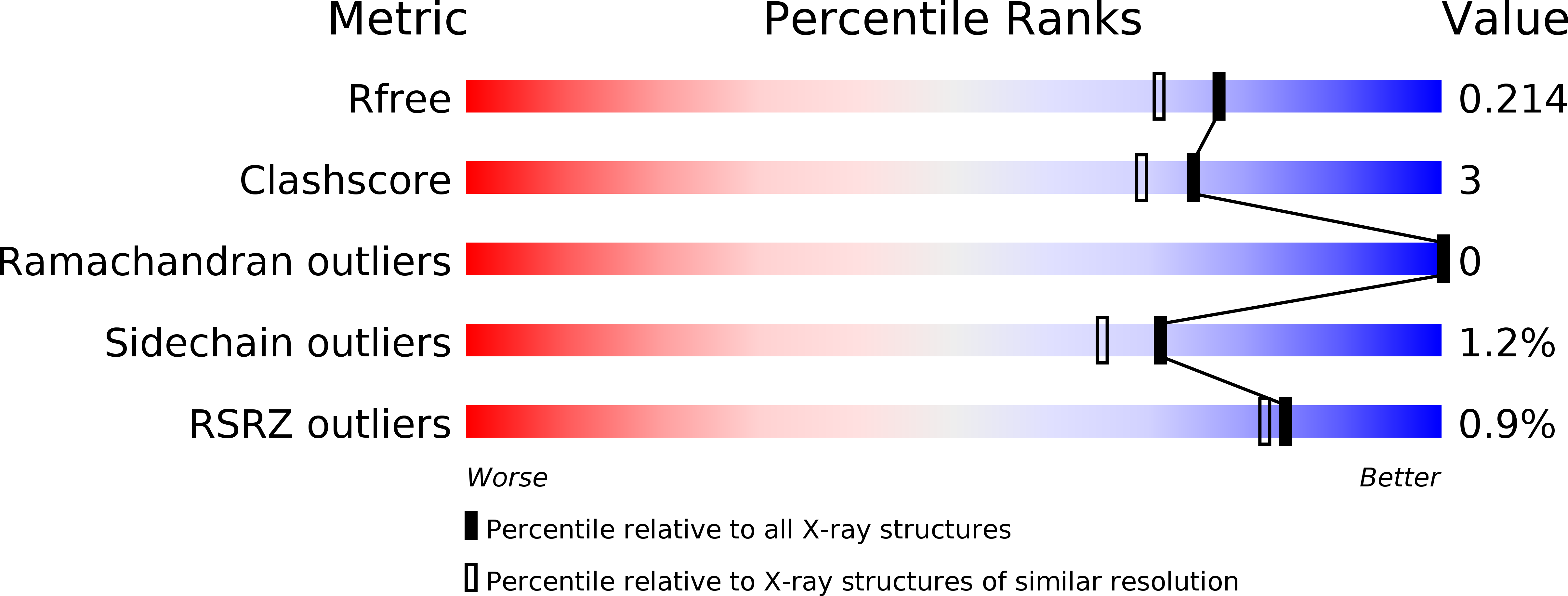
Roles of charged residues in pH-dependent redox properties of cytochrome c3 from Desulfovibrio vulgaris Miyazaki F
Yahata, N., Ozawa, K., Tomimoto, Y., Morita, K., Komori, H., Ogata, H., Higuchi, Y., Akutsu, H.(2006) Biophysics (Nagoya-shi) 2: 45-56