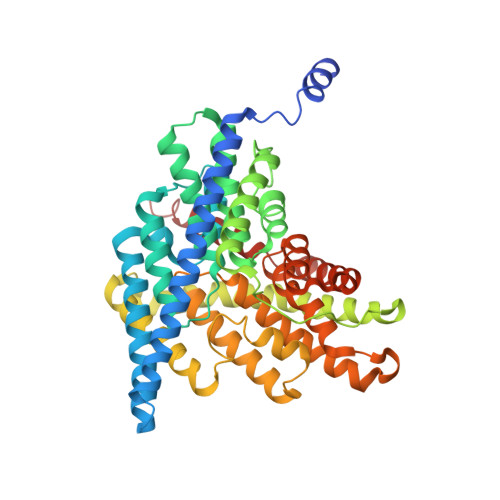
Separate ion pathways in a Cl-/H+ exchanger
Accardi, A., Walden, M.P., Nguitragool, W., Jayaram, H., Williams, C., Miller, C.(2005) J Gen Physiol 126: 563-570
- PubMed: 16316975
- DOI: https://doi.org/10.1085/jgp.200509417
- Primary Citation of Related Structures:
2FEC, 2FED, 2FEE - PubMed Abstract:
CLC-ec1 is a prokaryotic CLC-type Cl(-)/H+ exchange transporter. Little is known about the mechanism of H+ coupling to Cl-. A critical glutamate residue, E148, was previously shown to be required for Cl(-)/H+ exchange by mediating proton transfer between the protein and the extracellular solution. To test whether an analogous H+ acceptor exists near the intracellular side of the protein, we performed a mutagenesis scan of inward-facing carboxyl-bearing residues and identified E203 as the unique residue whose neutralization abolishes H+ coupling to Cl- transport. Glutamate at this position is strictly conserved in all known CLCs of the transporter subclass, while valine is always found here in CLC channels. The x-ray crystal structure of the E203Q mutant is similar to that of the wild-type protein. Cl- transport rate in E203Q is inhibited at neutral pH, and the double mutant, E148A/E203Q, shows maximal Cl- transport, independent of pH, as does the single mutant E148A. The results argue that substrate exchange by CLC-ec1 involves two separate but partially overlapping permeation pathways, one for Cl- and one for H+. These pathways are congruent from the protein's extracellular surface to E148, and they diverge beyond this point toward the intracellular side. This picture demands a transport mechanism fundamentally different from familiar alternating-access schemes.
Organizational Affiliation:
Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, MA 02454, USA.