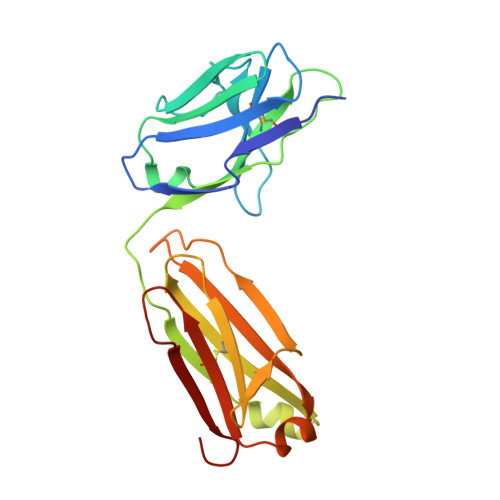
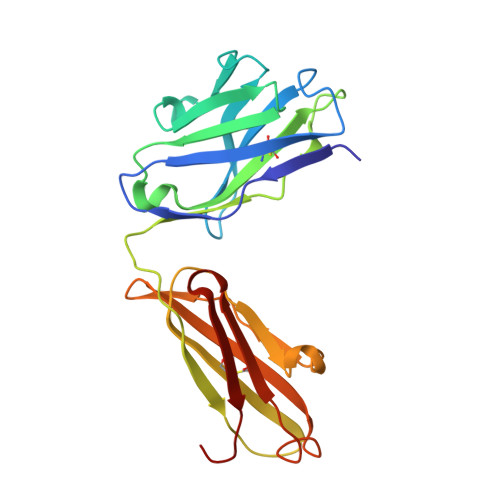
An anti-urokinase plasminogen activator receptor (uPAR) antibody: crystal structure and binding epitope
Li, Y., Parry, G., Chen, L., Callahan, J.A., Shaw, D.E., Meehan, E.J., Mazar, A.P., Huang, M.(2007) J Mol Biol 365: 1117-1129
- PubMed: 17101149
- DOI: https://doi.org/10.1016/j.jmb.2006.10.059
- Primary Citation of Related Structures:
2FAT - PubMed Abstract:
Human urokinase-type plasminogen activator receptor (uPAR/CD87) is expressed at the invasive interface of the tumor-stromal microenvironment in many human cancers and interacts with a wide array of extracellular molecules. An anti-uPAR antibody (ATN615) was prepared using hybridoma technology. This antibody binds to uPAR in vitro with high affinity (K(d) approximately 1 nM) and does not interfere with uPA binding to uPAR. Here we report the crystal structure of the Fab fragment of ATN615 at 1.77 A and the analysis of ATN615-suPAR-ATF structure that was previously determined, emphasizing the ATN615-suPAR interaction. The complementarity determining regions (CDRs) of ATN615 consist of a high percentage of aromatic residues, and form a relatively flat and undulating surface. The ATN615 Fab fragment recognizes domain 3 of suPAR. The antibody-antigen recognition involves 11 suPAR residues and 12 Fab residues from five CDRs. Structural data suggest that Pro188, Asn190, Gly191, and Arg192 residues of uPAR are the key residues for the antibody recognition, while Pro189 and Arg192 render specificity of ATN615 for human uPAR. Interestingly, this antibody-antigen interface has a small contact area, mainly polar interaction with little hydrophobic character, yet has high binding strength. Furthermore, several solvent molecules (assigned as polyethylene glycols) were clearly visible in the binding interface between antibody and antigen, suggesting that solvent molecules may be important for the maximal binding between suPAR and ATN615 Fab. ATN615 undergoes small but noticeable changes in its CDR region upon antigen binding.
Organizational Affiliation:
State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, 155 Yang Qiao Xi Lu, Fuzhou 350002, People's Republic of China.