Structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Miyazaki F)
Osuka, H., Suto, K., Sato, M., Shibata, N., Kitamura, M., Morimoto, Y., Ozawa, K., Akutsu, H., Higuchi, Y., Yasuoka, N.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| High-molecular-weight cytochrome c | 556 | Nitratidesulfovibrio vulgaris str. 'Miyazaki F | Mutation(s): 0 | 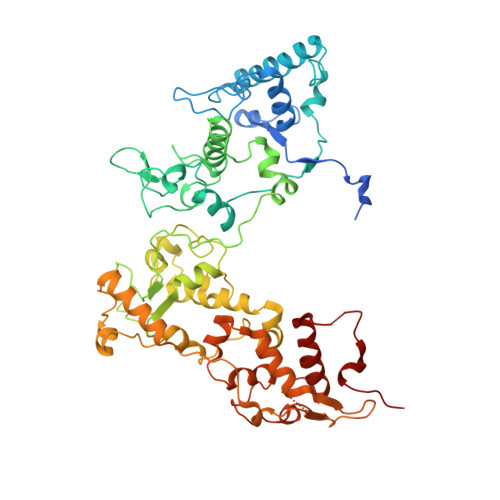 | |
UniProt | |||||
Find proteins for Q8VUI3 (Desulfovibrio vulgaris) Explore Q8VUI3 Go to UniProtKB: Q8VUI3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8VUI3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | J [auth A] K [auth A] L [auth A] M [auth A] N [auth A] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| ZN Query on ZN | B [auth A], C [auth A], D [auth A], E [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| NA Query on NA | F [auth A], G [auth A], H [auth A], I [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.09 | α = 90 |
| b = 84.07 | β = 90 |
| c = 142.67 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| MAR345 | data collection |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| CNS | phasing |