Exploring Chromophore-Protein Interactions in Fluorescent Protein Cmfp512 from Cerianthus Membranaceus: X-Ray Structure Analysis and Optical Spectroscopy.
Nienhaus, K., Renzi, F., Vallone, B., Wiedenmann, J., Nienhaus, G.U.(2006) Biochemistry 45: 12492
- PubMed: 17059211
- DOI: https://doi.org/10.1021/bi060885c
- Primary Citation of Related Structures:
2C9J - PubMed Abstract:
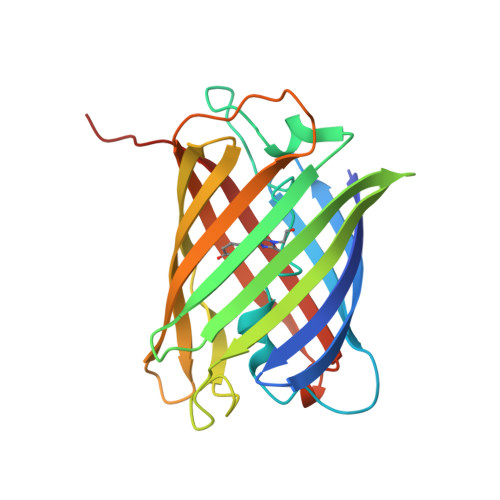
Autofluorescent proteins of the GFP family all share the same three-dimensional beta-can fold; yet they exhibit widely different optical properties, arising either from chemical modification of the chromophore itself or from specific interactions of the chromophore with the surrounding protein moiety. Here we present a structural and spectroscopic characterization of the green fluorescent protein cmFP512 from Cerianthus membranaceus, a nonbioluminescent, azooxanthellate cnidarian, which has only approximately 22% sequence identity with Aequorea victoria GFP. The X-ray structure, obtained by molecular replacement at a resolution of 1. 35 A, shows the chromophore, formed from the tripeptide Gln-Tyr-Gly, in a hydrogen-bonded cage in the center of an 11-stranded beta-barrel, tightly restrained by adjacent residues and structural water molecules. It exists in a neutral (A) and an anionic (B) species, with absorption/emission maxima at 392/460 (pH 5) and 503/512 nm (pH 7). Their fractional populations and peak positions depend sensitively on pH, reflecting protonation of groups adjacent to the chromophore. The pH dependence of the spectra is explained by a protonation mechanism involving a hydrogen-bonded cluster of charged/polar groups. Cryospectroscopy at 12 K was also performed to analyze the vibronic coupling of the electronic transitions.
Organizational Affiliation:
Department of Biophysics, University of Ulm, 89069 Ulm, Germany.