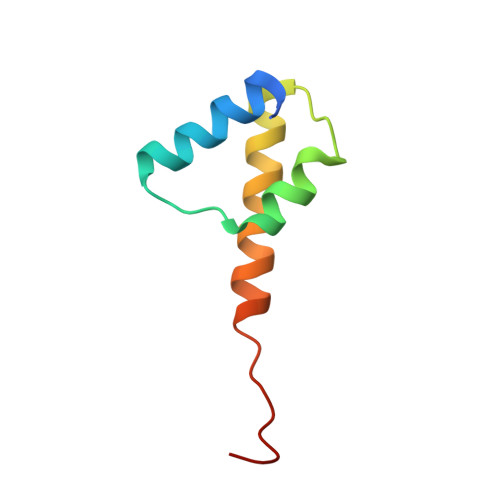
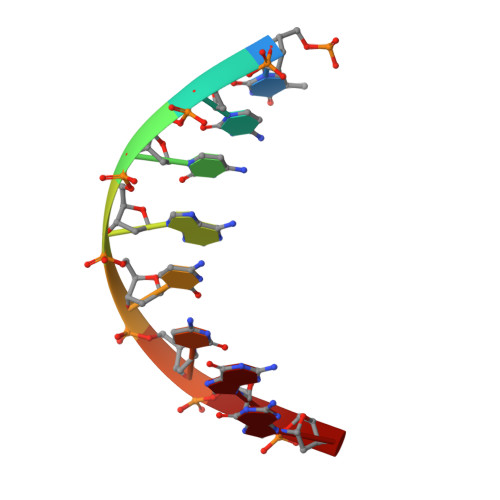
Structural Basis for Membrane Anchorage of Viral Phi 29 DNA During Replication.
Albert, A., Munoz-Espin, D., Jimenez, M., Asensio, J.L., Hermoso, J.A., Salas, M., Meijer, W.J.J.(2005) J Biol Chem 280: 42486
- PubMed: 16275651
- DOI: https://doi.org/10.1074/jbc.C500429200
- Primary Citation of Related Structures:
2C5R - PubMed Abstract:
Prokaryotic DNA replication is compartmentalized at the cellular membrane. Functional and biochemical studies showed that the Bacillus subtilis phage 29-encoded membrane protein p16.7 is directly involved in the organization of membrane-associated viral DNA replication. The structure of the functional domain of p16.7 in complex with DNA, presented here, reveals the multimerization mode of the protein and provides insights in the organization of the phage genome at the membrane of the infected cell.
Organizational Affiliation:
Grupo de Cristalografía Macromolecular y Biología Estructural, Instituto de Química-Física "Rocasolano", CSIC, Serrano 119, 28006 Madrid, Spain. xalbert@iqfr.csic.es