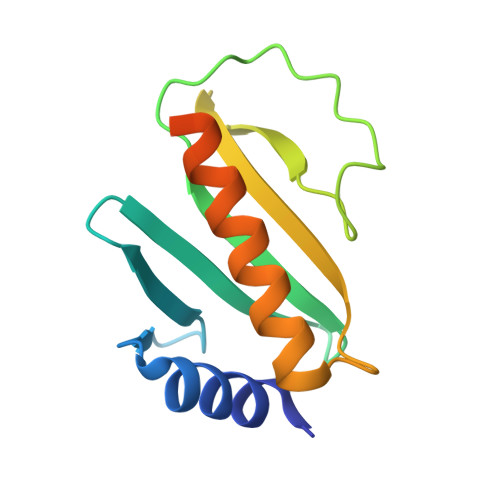
Crystal Structure of the Yersinia Enterocolitica Type III Secretion Chaperone Syct
Locher, M., Lehnert, B., Krauss, K., Heesemann, J., Groll, M., Wilharm, G.(2005) J Biol Chem 280: 31149
- PubMed: 16000312
- DOI: https://doi.org/10.1074/jbc.M500603200
- Primary Citation of Related Structures:
2BHO - PubMed Abstract:
Several Gram-negative pathogens deploy type III secretion systems (TTSSs) as molecular syringes to inject effector proteins into host cells. Prior to secretion, some of these effectors are accompanied by specific type III secretion chaperones. The Yersinia enterocolitica TTSS chaperone SycT escorts the effector YopT, a cysteine protease that inactivates the small GTPase RhoA of targeted host cells. We solved the crystal structure of SycT at 2.5 angstroms resolution. Despite limited sequence similarity among TTSS chaperones, the SycT structure revealed a global fold similar to that exhibited by other structurally solved TTSS chaperones. The dimerization domain of SycT, however, differed from that of all other known TTSS chaperone structures. Thus, the dimerization domain of TTSS chaperones does not likely serve as a general recognition pattern for downstream processing of effector/chaperone complexes. Yersinia Yop effectors are bound to their specific Syc chaperones close to the Yop N termini, distinct from their catalytic domains. Here, we showed that the catalytically inactive YopT(C139S) is reduced in its ability to bind SycT, suggesting an ancillary interaction between YopT and SycT. This interaction could maintain the protease inactive prior to secretion or could influence the secretion competence and folding of YopT.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Am Klopferspitz 18a, D-82152 Martinsried, Germany.