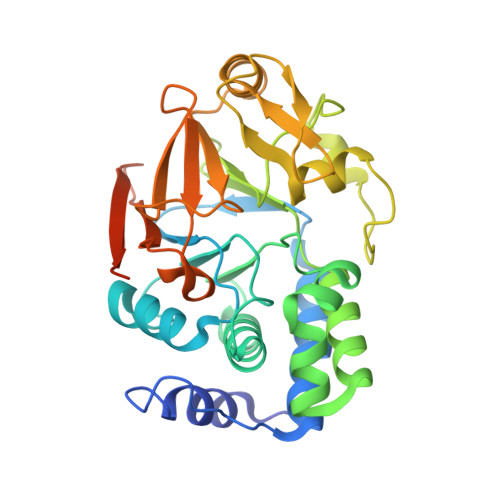
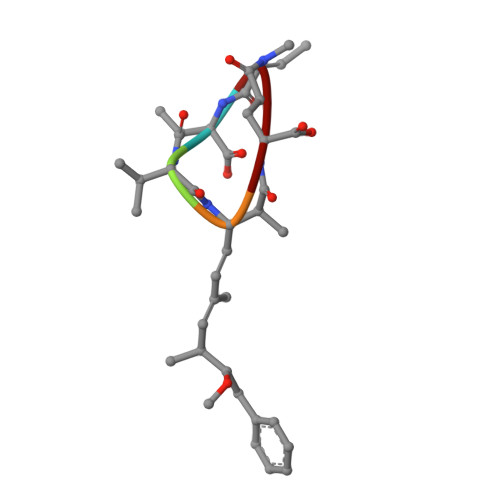
Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
Maynes, J.T., Luu, H.A., Cherney, M.M., Andersen, R.J., Williams, D., Holmes, C.F., James, M.N.(2006) J Mol Biol 356: 111-120
- PubMed: 16343532
- DOI: https://doi.org/10.1016/j.jmb.2005.11.019
- Primary Citation of Related Structures:
2BCD, 2BDX - PubMed Abstract:
The microcystins and nodularins are tumour promoting hepatotoxins that are responsible for global adverse human health effects and wildlife fatalities in countries where drinking water supplies contain cyanobacteria. The toxins function by inhibiting broad specificity Ser/Thr protein phosphatases in the host cells, thereby disrupting signal transduction pathways. A previous crystal structure of a microcystin bound to the catalytic subunit of protein phosphatase-1 (PP-1c) showed distinct changes in the active site region when compared with protein phosphatase-1 structures bound to other toxins. We have elucidated the crystal structures of the cyanotoxins, motuporin (nodularin-V) and dihydromicrocystin-LA bound to human protein phosphatase-1c (gamma isoform). The atomic structures of these complexes reveal the structural basis for inhibition of protein phosphatases by these toxins. Comparisons of the structures of the cyanobacterial toxin:phosphatase complexes explain the biochemical mechanism by which microcystins but not nodularins permanently modify their protein phosphatase targets by covalent addition to an active site cysteine residue.
Organizational Affiliation:
Canadian Institutes of Health Research, Group in Protein Structure and Function Department of Biochemistry, Faculty of Medicine, University of Alberta, Edmonton, Alta, Canada T6G 2H7.