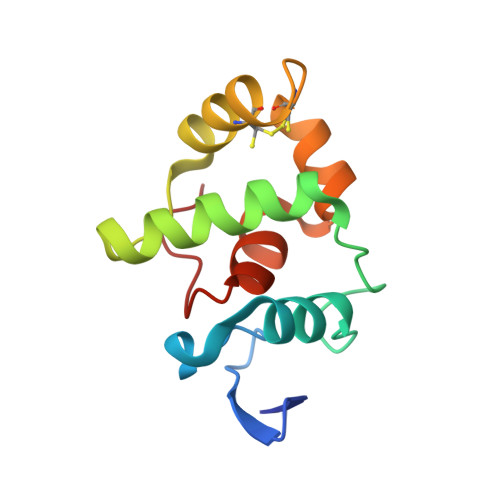
X-ray structure of the gamma-subunit of a dissimilatory sulfite reductase: Fixed and flexible C-terminal arms.
Mander, G.J., Weiss, M.S., Hedderich, R., Kahnt, J., Ermler, U., Warkentin, E.(2005) FEBS Lett 579: 4600-4604
- PubMed: 16098517
- DOI: https://doi.org/10.1016/j.febslet.2005.07.029
- Primary Citation of Related Structures:
2A5W - PubMed Abstract:
The X-ray structure of the gamma-subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus was determined at 1.12 and 2.1A resolution, in the two crystal forms named DsrC(nat) and DsrC(ox) the latter being cocrystallized with the oxidizing agent tert-butyl hydroperoxide. The fold corresponds to that of the homologous protein from Pyrobaculum aerophilum but is significantly more compact. The most interesting, highly conserved C-terminal arm adopts a well-defined conformation in A. fulgidus DsrC in contrast to the completely disordered conformation in P. aerophilum DsrC. The functional relevance of both conformations and of a potentially redox-active disulfide bond between the strictly invariant Cys103 and Cys114 are discussed.
Organizational Affiliation:
Max-Planck-Institut für terrestrische Mikrobiologie, Karl-von-Frisch-Str., D-35043 Marburg, Germany.