Structure and mechanism of the reversible photoswitch of a fluorescent protein
Andresen, M., Wahl, M.C., Stiel, A.C., Graeter, F., Schaefer, L., Trowitzsch, S., Weber, G., Eggeling, C., Grubmueller, H., Hell, S.W., Jakobs, S.(2005) Proc Natl Acad Sci U S A 102: 13070-13074
- PubMed: 16135569
- DOI: https://doi.org/10.1073/pnas.0502772102
- Primary Citation of Related Structures:
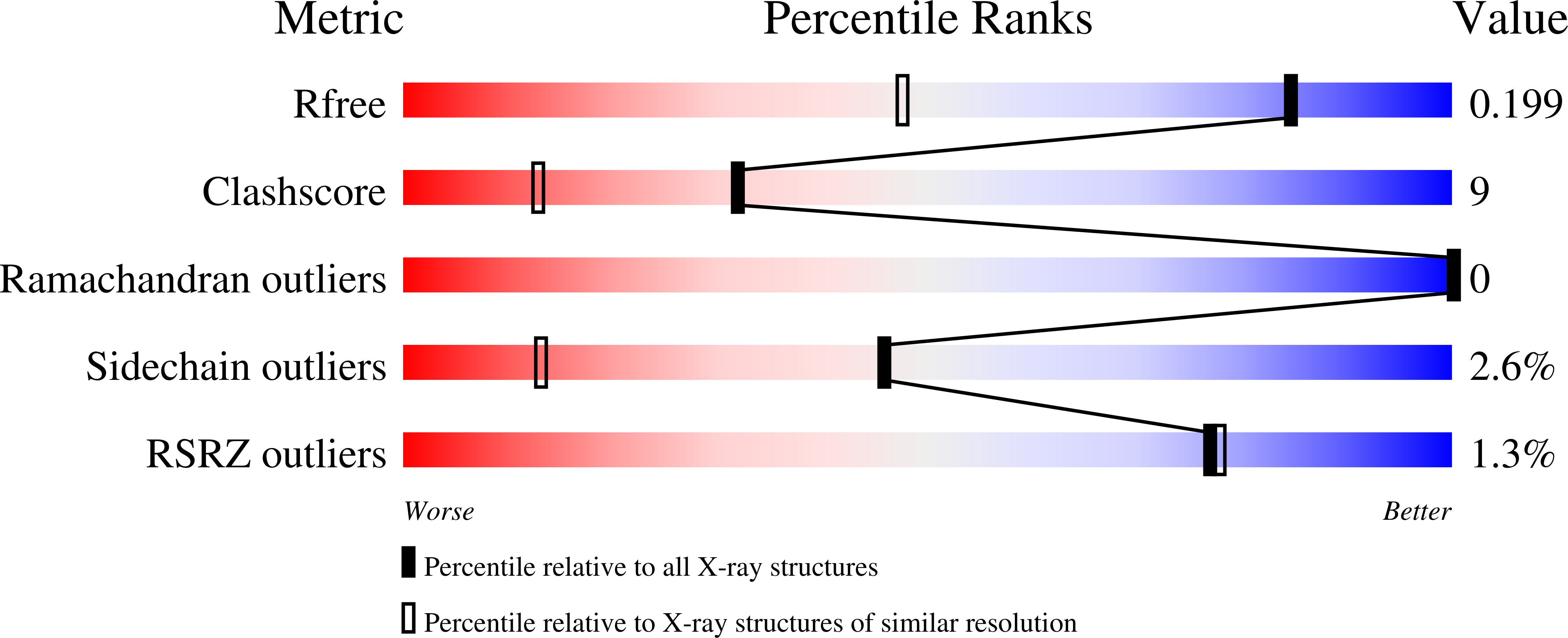
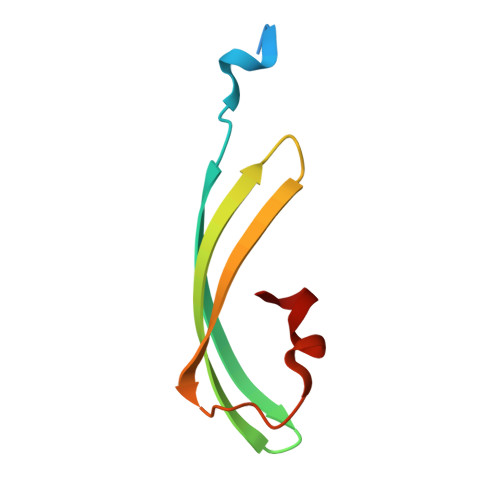
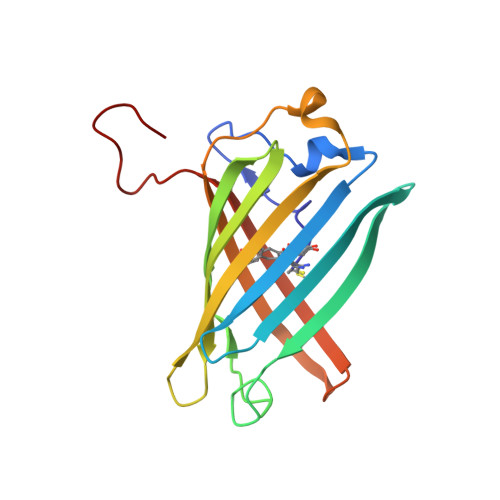
2A50, 2A52, 2A53, 2A54, 2A56 - PubMed Abstract:
Proteins that can be reversibly photoswitched between a fluorescent and a nonfluorescent state bear enormous potential in diverse fields, such as data storage, in vivo protein tracking, and subdiffraction resolution light microscopy. However, these proteins could hitherto not live up to their full potential because the molecular switching mechanism is not resolved. Here, we clarify the molecular photoswitching mechanism of asFP595, a green fluorescent protein (GFP)-like protein that can be transferred from a nonfluorescent "off" to a fluorescent "on" state and back again, by green and blue light, respectively. To this end, we establish reversible photoswitching of fluorescence in whole protein crystals and show that the switching kinetics in the crystal is identical with that in solution. Subsequent x-ray analysis demonstrated that upon the absorption of a green photon, the chromophore isomerizes from a trans (off) to a cis (on) state. Molecular dynamics calculations suggest that isomerization occurs through a bottom hula twist mechanism with concomitant rotation of both bonds of the chromophoric methine ring bridge. This insight into the switching mechanism should facilitate the targeted design of photoswitchable proteins. Reversible photoswitching of the protein chromophore system within intact crystals also constitutes a step toward the use of fluorescent proteins in three-dimensional data recording.
Organizational Affiliation:
Department of NanoBiophotonics, Theoretical and Computational Biophysics, and X-Ray Crystallography, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.