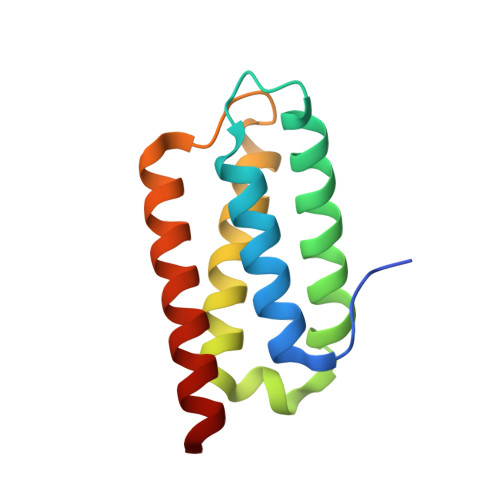
Crystal Structure of the Psb27 Assembly Factor at 1.6A: Implications for Binding to Photosystem II.
Michoux, F., Takasaka, K., Boehm, M., Komenda, J., Nixon, P.J., Murray, J.W.(2012) Photosynth Res 110: 169
- PubMed: 22193820
- DOI: https://doi.org/10.1007/s11120-011-9712-7
- Primary Citation of Related Structures:
2Y6X - PubMed Abstract:
The biogenesis and oxygen-evolving activity of cyanobacterial Photosystem II (PSII) is dependent on a number of accessory proteins not found in the crystallised dimeric complex. These include Psb27, a small lipoprotein attached to the lumenal side of PSII, which has been assigned a role in regulating the assembly of the Mn(4)Ca cluster catalysing water oxidation. To gain a better understanding of Psb27, we have determined in this study the crystal structure of the soluble domain of Psb27 from Thermosynechococcus elongatus to a resolution of 1.6 Å. The structure is a four-helix bundle, similar to the recently published solution structures of Psb27 from Synechocystis PCC 6803 obtained by nuclear magnetic resonance (NMR) spectroscopy. Importantly, the crystal structure presented here helps us resolve the differences between the NMR-derived structural models. Potential binding sites for Psb27 within PSII are discussed in light of recent biochemical data in the literature.
Organizational Affiliation:
Division of Molecular Biosciences, Imperial College London, Wolfson Biochemistry Building, South Kensington Campus, London, SW7 2AZ, UK. franck.michoux@imperial.ac.uk