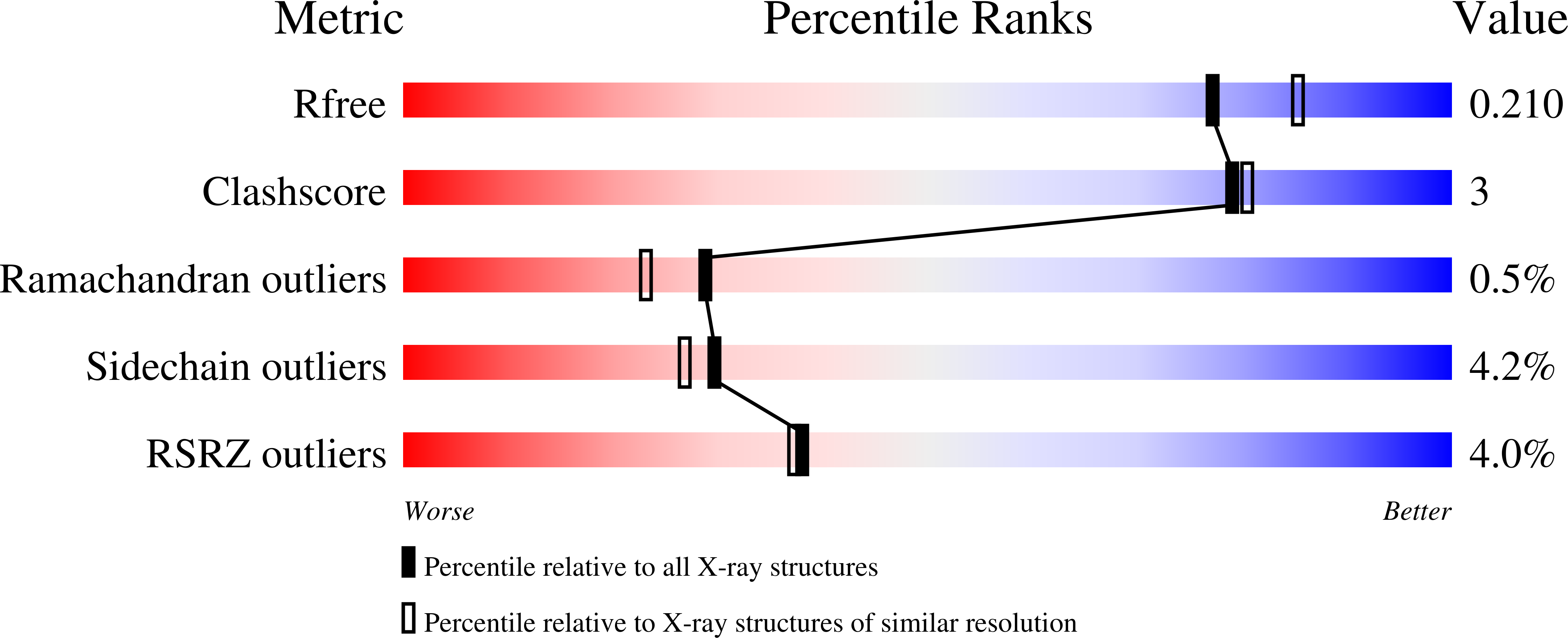
Novel Antiviral C5-Substituted Pyrimidine Acyclic Nucleoside Phosphonates Selected as Human Thymidylate Kinase Substrates.
Topalis, D., Pradere, U., Roy, V., Caillat, C., Azzouzi, A., Broggi, J., Snoeck, R., Andrei, G., Lin, J., Eriksson, S., Alexandre, J.A.C., El-Amri, C., Deville-Bonne, D., Meyer, P., Balzarini, J., Agrofoglio, L.A.(2011) J Med Chem 54: 222
- PubMed: 21128666
- DOI: https://doi.org/10.1021/jm1011462
- Primary Citation of Related Structures:
2XX3 - PubMed Abstract:
Acyclic nucleoside phosphonates (ANPs) are at the cornerstone of DNA virus and retrovirus therapies. They reach their target, the viral DNA polymerase, after two phosphorylation steps catalyzed by cellular kinases. New pyrimidine ANPs have been synthesized with unsaturated acyclic side chains (prop-2-enyl-, but-2-enyl-, pent-2-enyl-) and different substituents at the C5 position of the uracil nucleobase. Several derivatives in the but-2-enyl- series 9d and 9e, with (E) but not with (Z) configuration, were efficient substrates for human thymidine monophosphate (TMP) kinase, but not for uridine monophosphate-cytosine monophosphate (UMP-CMP) kinase, which is in contrast to cidofovir. Human TMP kinase was successfully crystallized in a complex with phosphorylated (E)-thymidine-but-2-enyl phosphonate 9e and ADP. The bis-pivaloyloxymethyl (POM) esters of (E)-9d and (E)-9e were synthesized and shown to exert activity against herpes virus in vitro (IC(50) = 3 μM) and against varicella zoster virus in vitro (IC(50) = 0.19 μM), in contrast to the corresponding inactive (Z) derivatives. Thus, their antiviral activity correlates with their ability to act as thymidylate kinase substrates.
Organizational Affiliation:
Groupe d'Enzymologie Moléculaire et Fonctionnelle, UR4-UPMC, Université Pierre et Marie Curie, Sorbonne Universités, Paris, France.