The K5 Lyase Kfla Combines a Viral Tail Spike Structure with a Bacterial Polysaccharide Lyase Mechanism.
Thompson, J.E., Pourhossein, M., Waterhouse, A., Hudson, T., Goldrick, M., Derrick, J.P., Roberts, I.S.(2010) J Biol Chem 285: 23963
- PubMed: 20519506
- DOI: https://doi.org/10.1074/jbc.M110.127571
- Primary Citation of Related Structures:
2X3H - PubMed Abstract:
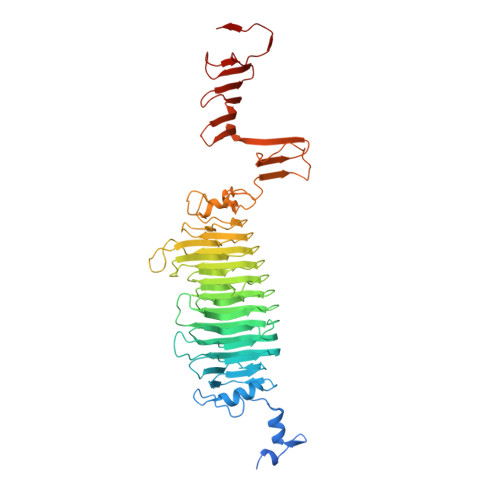
K5 lyase A (KflA) is a tail spike protein (TSP) encoded by a K5A coliphage, which cleaves K5 capsular polysaccharide, a glycosaminoglycan with the repeat unit [-4)-betaGlcA-(1,4)- alphaGlcNAc(1-], displayed on the surface of Escherichia coli K5 strains. The crystal structure of KflA reveals a trimeric arrangement, with each monomer containing a right-handed, single-stranded parallel beta-helix domain. Stable trimer formation by the intertwining of strands in the C-terminal domain, followed by proteolytic maturation, is likely to be catalyzed by an autochaperone as described for K1F endosialidase. The structure of KflA represents the first bacteriophage tail spike protein combining polysaccharide lyase activity with a single-stranded parallel beta-helix fold. We propose a catalytic site and mechanism representing convergence with the syn-beta-elimination site of heparinase II from Pedobacter heparinus.
Organizational Affiliation:
Faculty of Life Sciences, University of Manchester, Manchester M13 9PT, United Kingdom.