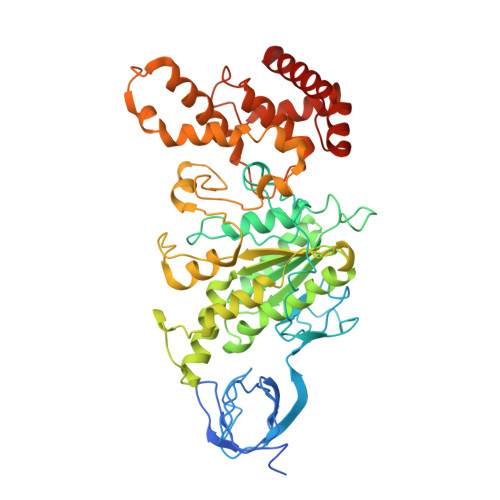
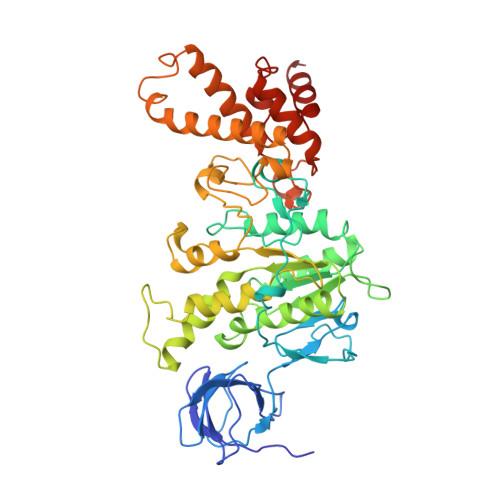
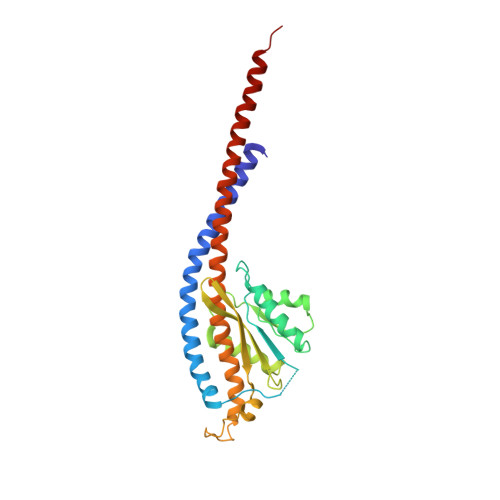
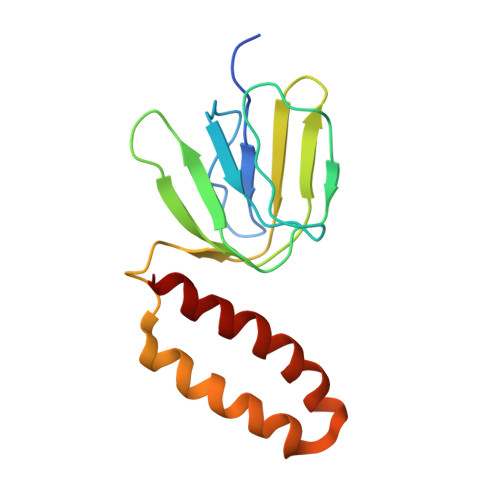
Crystal Structure of the Mg.Adp-Inhibited State of the Yeast F1C10-ATP Synthase.
Dautant, A., Velours, J., Giraud, M.(2010) J Biol Chem 285: 29502
- PubMed: 20610387
- DOI: https://doi.org/10.1074/jbc.M110.124529
- Primary Citation of Related Structures:
2WPD - PubMed Abstract:
The F(1)c(10) subcomplex of the yeast F(1)F(0)-ATP synthase includes the membrane rotor part c(10)-ring linked to a catalytic head, (αβ)(3), by a central stalk, γδε. The Saccharomyces cerevisiae yF(1)c(10)·ADP subcomplex was crystallized in the presence of Mg·ADP, dicyclohexylcarbodiimide (DCCD), and azide. The structure was solved by molecular replacement using a high resolution model of the yeast F(1) and a bacterial c-ring model with 10 copies of the c-subunit. The structure refined to 3.43-Å resolution displays new features compared with the original yF(1)c(10) and with the yF(1) inhibited by adenylyl imidodiphosphate (AMP-PNP) (yF(1)(I-III)). An ADP molecule was bound in both β(DP) and β(TP) catalytic sites. The α(DP)-β(DP) pair is slightly open and resembles the novel conformation identified in yF(1), whereas the α(TP)-β(TP) pair is very closed and resembles more a DP pair. yF(1)c(10)·ADP provides a model of a new Mg·ADP-inhibited state of the yeast F(1). As for the original yF(1) and yF(1)c(10) structures, the foot of the central stalk is rotated by ∼40 ° with respect to bovine structures. The assembly of the F(1) central stalk with the F(0) c-ring rotor is mainly provided by electrostatic interactions. On the rotor ring, the essential cGlu(59) carboxylate group is surrounded by hydrophobic residues and is not involved in hydrogen bonding.
Organizational Affiliation:
Université Bordeaux 2, CNRS, Institut de Biochimie et Génétique Cellulaires, 1 rue Camille Saint-Saëns, 33077 Bordeaux Cedex, France. A.Dautant@ibgc.cnrs.fr