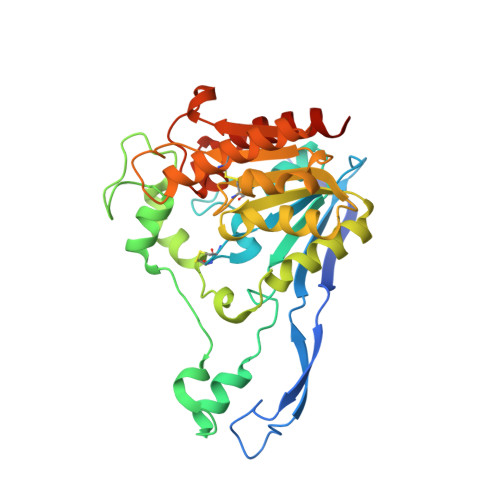
The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Bourne, P.C., Isupov, M.N., Littlechild, J.A.(2000) Structure 8: 143
- PubMed: 10673440
- DOI: https://doi.org/10.1016/s0969-2126(00)00090-3
- Primary Citation of Related Structures:
1QLW, 2WKW - PubMed Abstract:
A novel bacterial esterase that cleaves esters on halogenated cyclic compounds has been isolated from an Alcaligenes species. This esterase 713 is encoded by a 1062 base pair gene. The presence of a leader sequence of 27 amino acids suggests that this enzyme is exported from the cytosol. Esterase 713 has been over-expressed in Agrobacterium without this leader sequence. Its amino acid sequence shows no significant homology to any known protein sequence.
Organizational Affiliation:
Schools of Chemistry and Biological Sciences, University of Exeter, Exeter, EX4 4QD, UK.