Crystal Structure of Human Pus10, a Novel Pseudouridine Synthase.
Mccleverty, C.J., Hornsby, M., Spraggon, G., Kreusch, A.(2007) J Mol Biol 373: 1243
- PubMed: 17900615
- DOI: https://doi.org/10.1016/j.jmb.2007.08.053
- Primary Citation of Related Structures:
2V9K - PubMed Abstract:
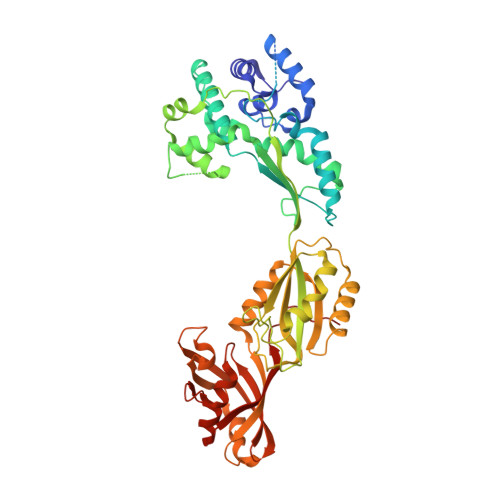
Pseudouridine (Psi) synthases catalyze the formation of one or more specific Psis in structured RNAs. Five families of Psi synthases have been characterized based on sequence homology. Pus10 has no significant sequence homology to these defined families and therefore represents a new family of Psi synthases. Initial characterization studies show that an archael Pus10 catalyzes the universally conserved Psi55 in tRNA. We present here the crystal structure of human Pus10 at 2.0 A resolution, which is the first structural description from this novel Psi synthase family. Pus10 is a crescent-shaped molecule with two domains, the universally conserved Psi synthase catalytic domain and a THUMP-containing domain, which is unique to the Pus10 family. Superposition of the catalytic domains of Pus10 and other Psi synthases identifies the full set of conserved Psi synthase active site residues indicating that Pus10 likely employs a similar catalytic mechanism to other Psi synthases. The Pus10 active site is located in a deep pocket of a basic cleft adjacent to flexible thumb and forefinger loops, which could provide further stabilization for binding the RNA substrate. Modeling studies demonstrate that the cleft between the catalytic and accessory domain is large enough and electrostatically compatible to accommodate an RNA stem and support the role of the N-terminal domain as an accessory RNA-binding domain.
Organizational Affiliation:
Genomics Institute of the Novartis Research Foundation, 10675 John Jay Hopkins Drive, San Diego, CA 92121, USA.