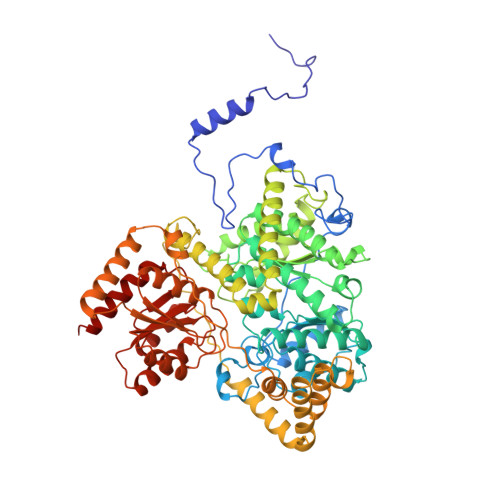
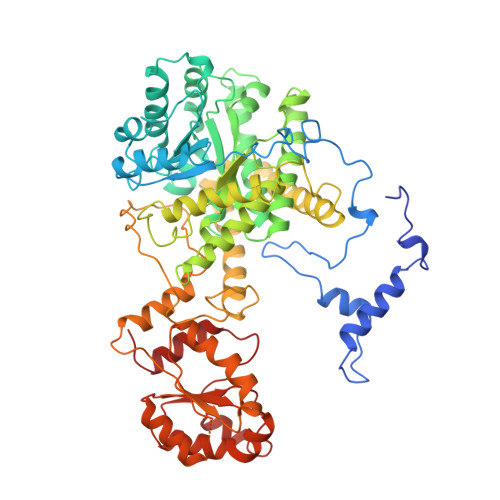
Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism.
Mancia, F., Evans, P.R.(1998) Structure 6: 711-720
- PubMed: 9655823
- DOI: https://doi.org/10.1016/s0969-2126(98)00073-2
- Primary Citation of Related Structures:
2REQ, 3REQ, 4REQ - PubMed Abstract:
Methylmalonyl CoA mutase catalyses the interconversion of succinyl CoA and methylmalonyl CoA via a free radical mechanism. The enzyme belongs to a family of enzymes that catalyse intramolecular rearrangement reactions in which a group and a hydrogen atom on adjacent carbons are exchanged. These enzymes use the cofactor adenosylcobalamin (coenzyme B12) which breaks to form an adenosyl radical, thus initiating the reaction. Determination of the structure of substrate-free methylmalonyl CoA mutase was initiated to provide further insight into the mechanism of radical formation.
Organizational Affiliation:
MRC Laboratory of Molecular Biology Hills Road, Cambridge, CB2 2QH, UK.