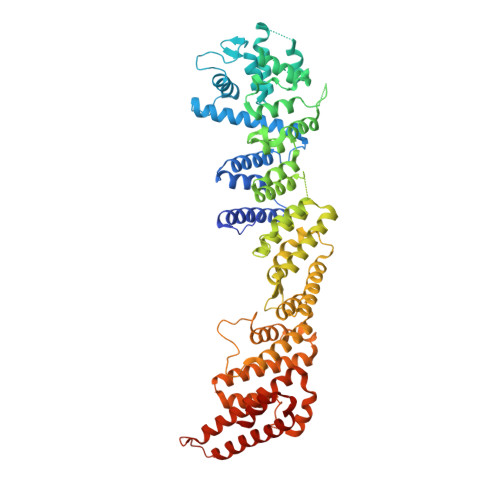
Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture
Jeudy, S., Schwartz, T.U.(2007) J Biol Chem 282: 34904
- PubMed: 17897938
- DOI: https://doi.org/10.1074/jbc.M705479200
- Primary Citation of Related Structures:
2QX5 - PubMed Abstract:
The nuclear pore complex (NPC) is an elaborate protein machine that mediates macromolecular transport across the nuclear envelope in all eukaryotes. The NPC is formed by nucleoporins that assemble in multiple copies around an 8-fold symmetry axis. Homology modeling suggests that most architectural nucleoporins are composed of simple beta-propeller and alpha-helical repeat domains. Here we present the crystal structure of Nic96, the Nup93 homolog in Saccharomyces cerevisiae, one of the major components of the NPC. This is the first structure of an alpha-helical nucleoporin domain. The protein folds into an elongated, mostly alpha-helical structure. Characteristically, non-canonical architectural features define the Nic96 structure. Sequence conservation among Nup93 homologs across all eukaryotes strongly suggests that the distinct topology is evolutionarily well maintained. We propose that the unique Nic96/Nup93 fold has a conserved function in all eukaryotes.
Organizational Affiliation:
Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.