Crystal Structure of Atp12 from Paracoccus Denitrificans
Ludlam, A.V., Brunzelle, J.S., Pribyl, T., Gatti, D.L., Ackerman, S.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP12 ATPase | 239 | Paracoccus denitrificans PD1222 | Mutation(s): 0 Gene Names: Pden_0792 | 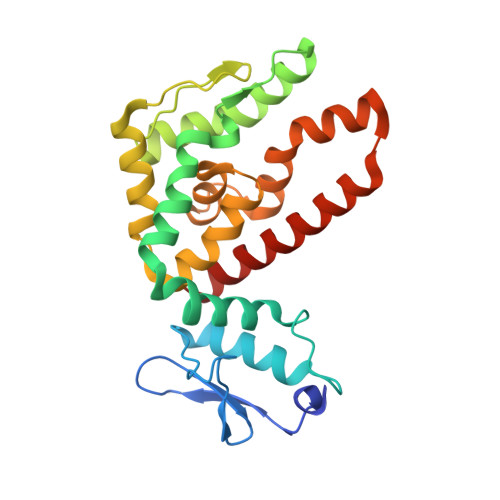 | |
UniProt | |||||
Find proteins for A1B060 (Paracoccus denitrificans (strain Pd 1222)) Explore A1B060 Go to UniProtKB: A1B060 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1B060 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.854 | α = 73.87 |
| b = 50.632 | β = 74.19 |
| c = 67.581 | γ = 89.78 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SOLVE | phasing |