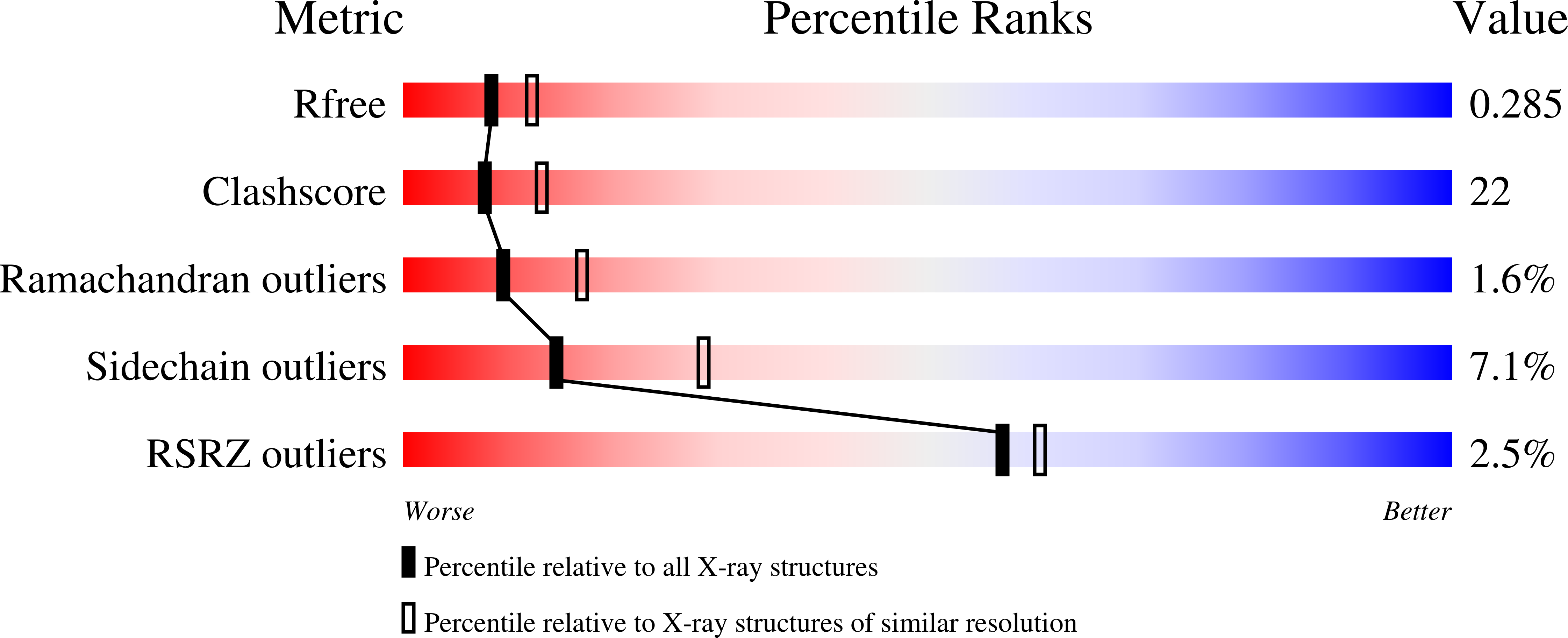
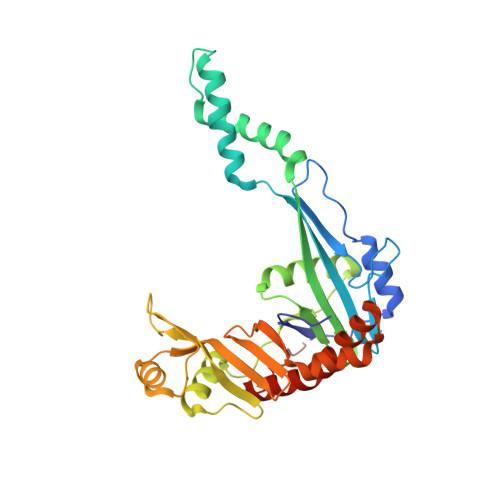
The recombination-associated protein RdgC adopts a novel toroidal architecture for DNA binding
Ha, J.Y., Kim, H.K., Kim, D.J., Kim, K.H., Oh, S.J., Lee, H.H., Yoon, H.J., Song, H.K., Suh, S.W.(2007) Nucleic Acids Res 35: 2671-2681
- PubMed: 17426134
- DOI: https://doi.org/10.1093/nar/gkm144
- Primary Citation of Related Structures:
2OWY - PubMed Abstract:
RecA plays a central role in the nonmutagenic repair of stalled replication forks in bacteria. RdgC, a recombination-associated DNA-binding protein, is a potential negative regulator of RecA function. Here, we have determined the crystal structure of RdgC from Pseudomonas aeruginosa. The J-shaped monomer has a unique fold and can be divided into three structural domains: tip domain, center domain and base domain. Two such monomers dimerize to form a ring-shaped molecule of approximate 2-fold symmetry. Of the two inter-subunit interfaces within the dimer, one interface ('interface A') between tip/center domains is more nonpolar than the other ('interface B') between base domains. The structure allows us to propose that the RdgC dimer binds dsDNA through the central hole of approximately 30 A diameter. The proposed model is supported by our DNA-binding assays coupled with mutagenesis, which indicate that the conserved positively charged residues on the protein surface around the central hole play important roles in DNA binding. The novel ring-shaped architecture of the RdgC dimer has significant implications for its role in homologous recombination.
Organizational Affiliation:
Department of Chemistry, College of Natural Sciences, Seoul National University, Seoul 151-742, Korea.