Fis targets assembly of the xis nucleoprotein filament to promote excisive recombination by phage lambda.
Papagiannis, C.V., Sam, M.D., Abbani, M.A., Yoo, D., Cascio, D., Clubb, R.T., Johnson, R.C.(2007) J Mol Biol 367: 328-343
- PubMed: 17275024
- DOI: https://doi.org/10.1016/j.jmb.2006.12.071
- Primary Citation of Related Structures:
2OG0 - PubMed Abstract:
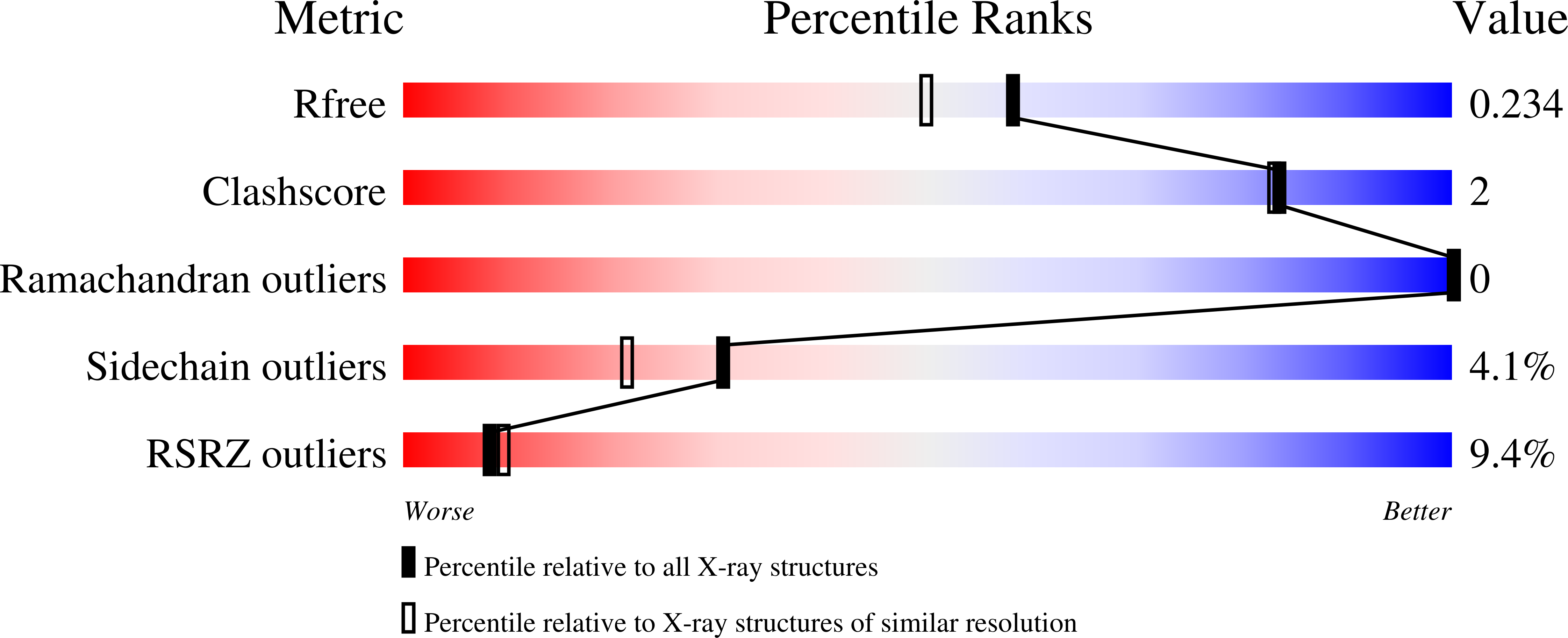
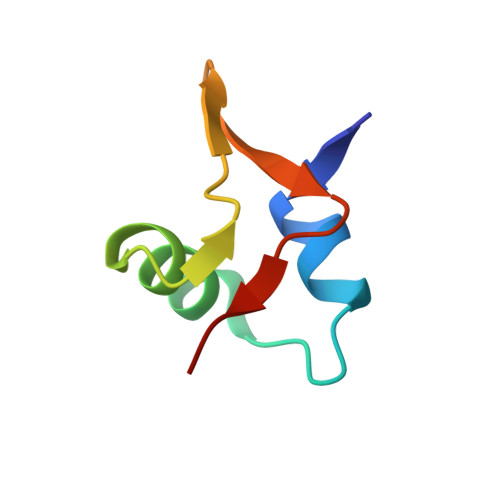
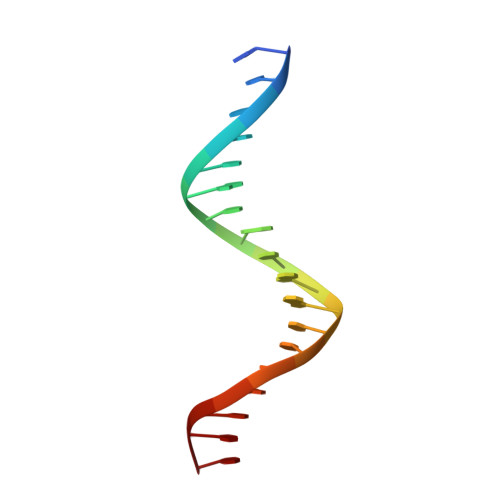
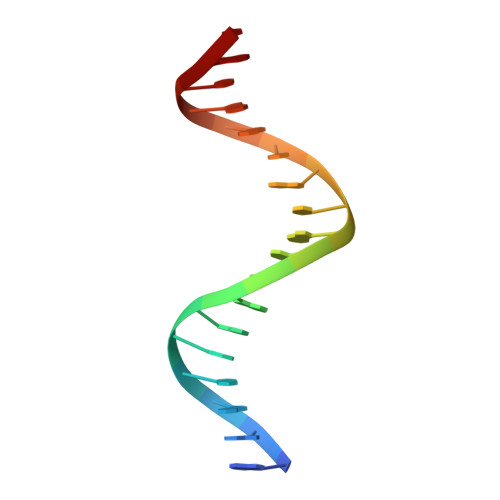
The phage-encoded Xis protein is the major determinant controlling the direction of recombination in phage lambda. Xis is a winged-helix DNA binding protein that cooperatively binds to the attR recombination site to generate a curved microfilament, which promotes assembly of the excisive intasome but inhibits formation of an integrative intasome. We find that lambda synthesizes surprisingly high levels of Xis immediately upon prophage induction when excision rates are maximal. However, because of its low sequence-specific binding activity, exemplified by a 1.9 A co-crystal structure of a non-specifically bound DNA complex, Xis is relatively ineffective at promoting excision in vivo in the absence of the host Fis protein. Fis binds to a segment in attR that almost entirely overlaps one of the Xis binding sites. Instead of sterically excluding Xis binding from this site, as has been previously believed, we show that Fis enhances binding of all three Xis protomers to generate the microfilament. A specific Fis-Xis interface is supported by the effects of mutations within each protein, and relaxed, but not completely sequence-neutral, binding by the central Xis protomer is supported by the effects of DNA mutations. We present a structural model for the 50 bp curved Fis-Xis cooperative complex that is assembled between the arm and core Int binding sites whose trajectory places constraints on models for the excisive intasome structure.
Organizational Affiliation:
Department of Biological Chemistry, David Geffen School of Medicine at UCLA, Los Angeles, CA 90095-1737, USA.