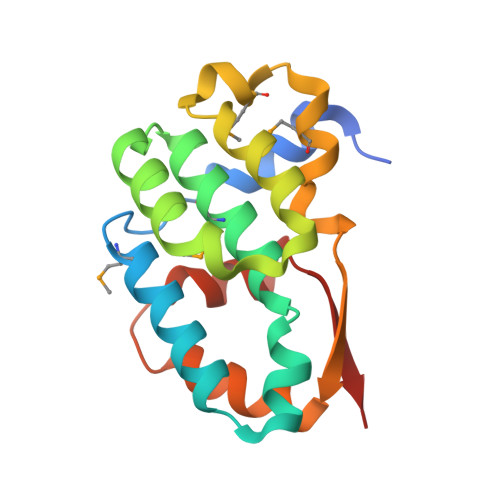
Structural analysis and dimerization potential of the human TAF5 subunit of TFIID.
Bhattacharya, S., Takada, S., Jacobson, R.H.(2007) Proc Natl Acad Sci U S A 104: 1189-1194
- PubMed: 17227857
- DOI: https://doi.org/10.1073/pnas.0610297104
- Primary Citation of Related Structures:
2NXP - PubMed Abstract:
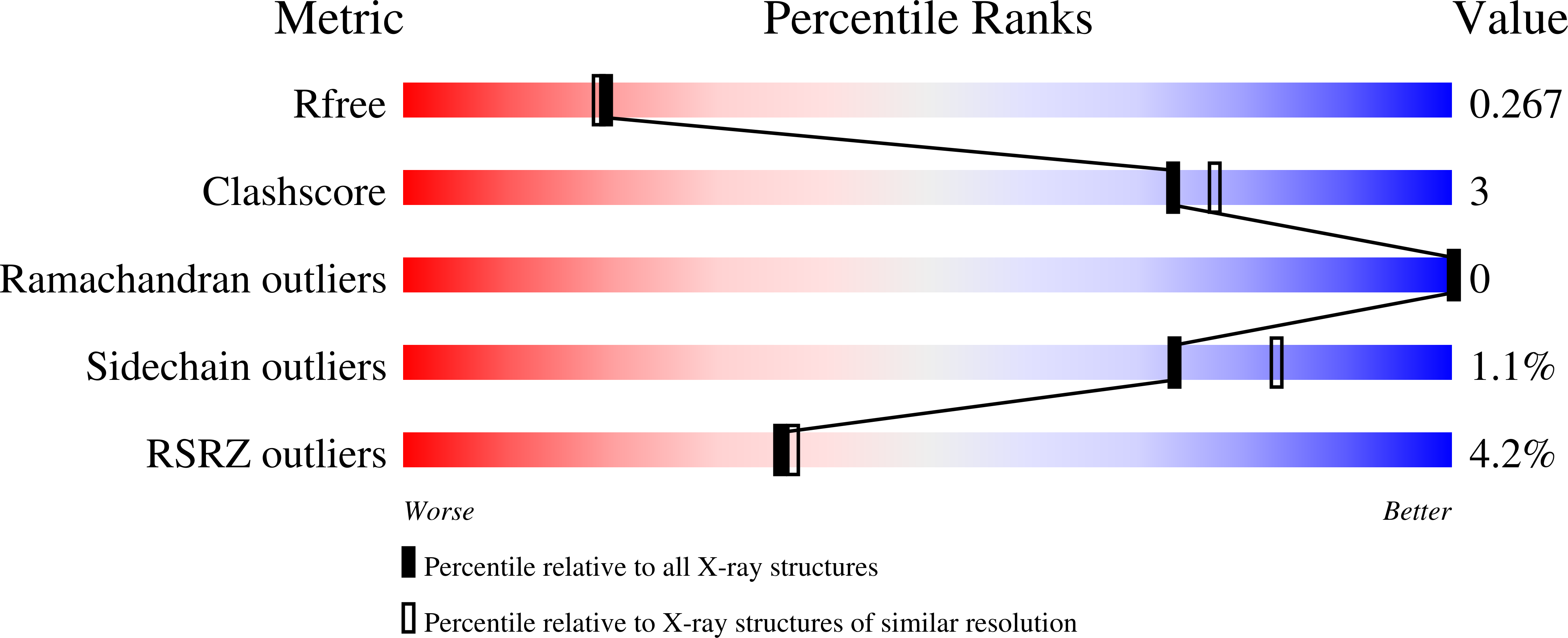
TFIID is an essential factor required for RNA polymerase II transcription but remains poorly understood because of its intrinsic complexity. Human TAF5, a 100-kDa subunit of general transcription factor TFIID, is an essential gene and plays a critical role in assembling the 1.2 MDa TFIID complex. We report here a structural analysis of the TAF5 protein. Our structure at 2.2-A resolution of the TAF5-NTD2 domain reveals an alpha-helical domain with distant structural similarity to RNA polymerase II CTD interacting factors. The TAF5-NTD2 domain contains several conserved clefts likely to be critical for TFIID complex assembly. Our biochemical analysis of the human TAF5 protein demonstrates the ability of the N-terminal half of the TAF5 gene to form a flexible, extended dimer, a key property required for the assembly of the TFIID complex.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Texas M. D. Anderson Cancer Center and the Program in Genes and Development at the University of Texas, Graduate School in Biochemical Sciences, Houston, TX 77030, USA.