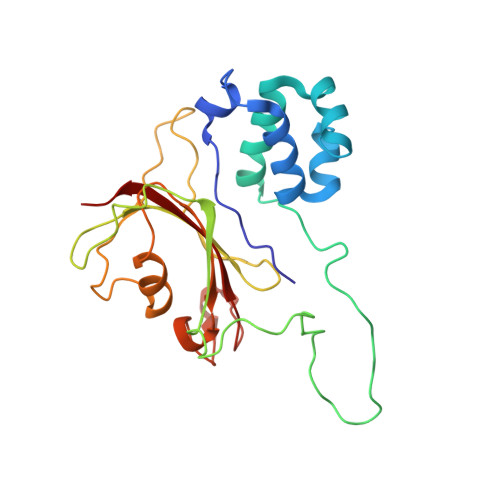
Structural basis of nucleic acid recognition by FK506-binding protein 25 (FKBP25), a nuclear immunophilin.
Prakash, A., Shin, J., Rajan, S., Yoon, H.S.(2016) Nucleic Acids Res 44: 2909-2925
- PubMed: 26762975
- DOI: https://doi.org/10.1093/nar/gkw001
- Primary Citation of Related Structures:
2MPH - PubMed Abstract:
The nuclear immunophilin FKBP25 interacts with chromatin-related proteins and transcription factors and is suggested to interact with nucleic acids. Currently the structural basis of nucleic acid binding by FKBP25 is unknown. Here we determined the nuclear magnetic resonance (NMR) solution structure of full-length human FKBP25 and studied its interaction with DNA. The FKBP25 structure revealed that the N-terminal helix-loop-helix (HLH) domain and C-terminal FK506-binding domain (FKBD) interact with each other and that both of the domains are involved in DNA binding. The HLH domain forms major-groove interactions and the basic FKBD loop cooperates to form interactions with an adjacent minor-groove of DNA. The FKBP25-DNA complex model, supported by NMR and mutational studies, provides structural and mechanistic insights into the nuclear immunophilin-mediated nucleic acid recognition.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551, Singapore.