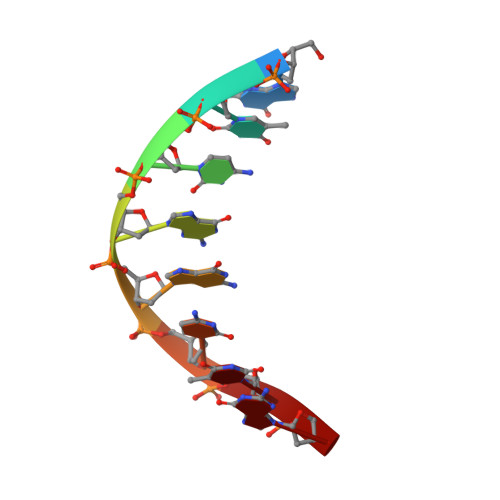
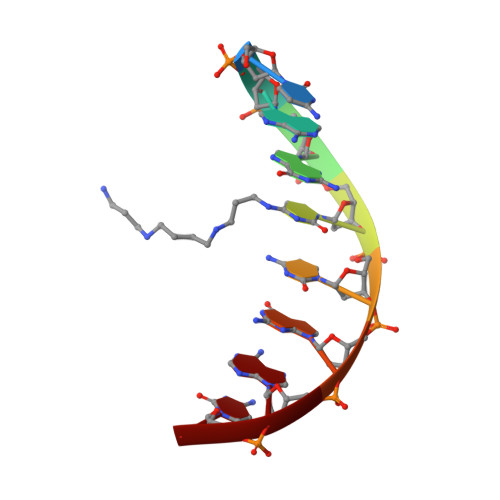
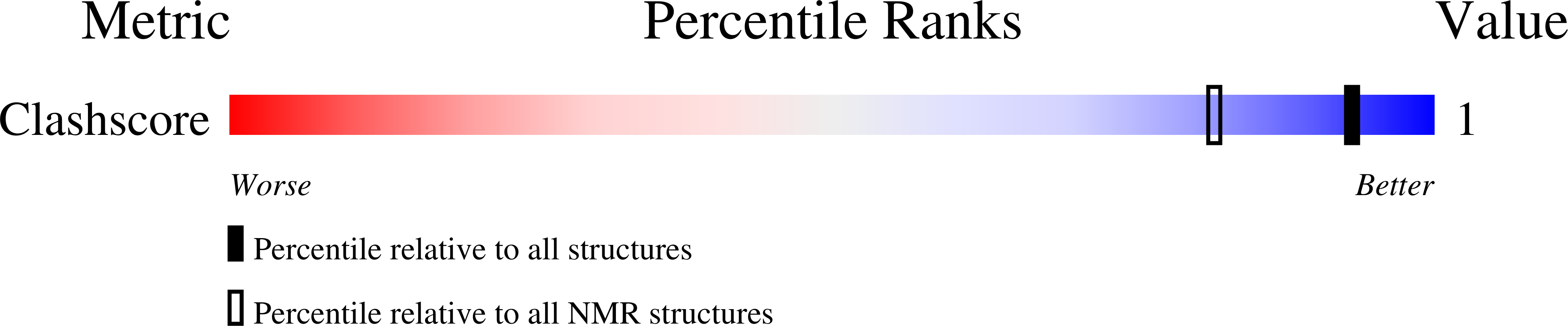Polyaminooligonucleotide: NMR structure of duplex DNA containing a nucleoside with spermine residue, N-[4,9,13-triazatridecan-1-yl]-2'-deoxycytidine.
Brzezinska, J., Gdaniec, Z., Popenda, L., Markiewicz, W.T.(2014) Biochim Biophys Acta 1840: 1163-1170
- PubMed: 24361616
- DOI: https://doi.org/10.1016/j.bbagen.2013.12.008
- Primary Citation of Related Structures:
2MCI, 2MCJ - PubMed Abstract:
The nature of the polyamine-DNA interactions at a molecular level is not clearly understood. In order to shed light on the binding preferences of polyamine with nucleic acids, the NMR solution structure of the DNA duplex containing covalently bound spermine was determined. The structure of 4-N-[4,9,13-triazatridecan-1-yl]-2'-deoxycytidine (dCSp) modified duplex was compared to the structure of the reference duplex. Both duplexes are regular right-handed helices with all attributes of the B-DNA form. The spermine chain which is located in a major groove and points toward the 3' end of the modified strand does not perturb the DNA structure. In our study the charged polyamine alkyl chain was found to interact with the DNA surface. In the majority of converged structures we identified the presumed hydrogen bonding interactions between O6 and N7 atoms of G4 and the first internal -NH2(+)- amino group. Additional interaction was found between the second internal -NH2(+)- amino group and the oxygen atom of the phosphate of C3 residue. The knowledge of the location and nature of a structure-specific binding site for spermine in DNA should be valuable in understanding gene expression and in the design of new therapeutic drugs.
Organizational Affiliation:
Institute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, PL-61704 Poznan, Poland.