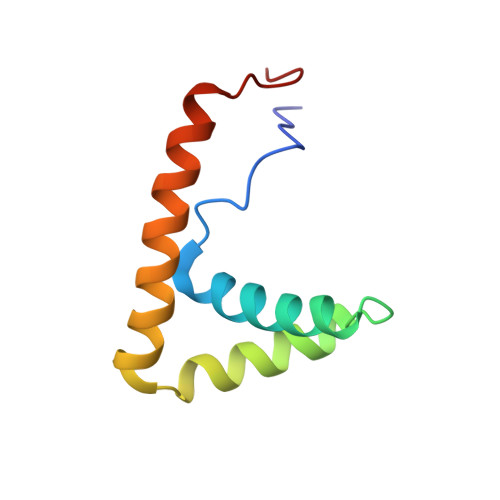
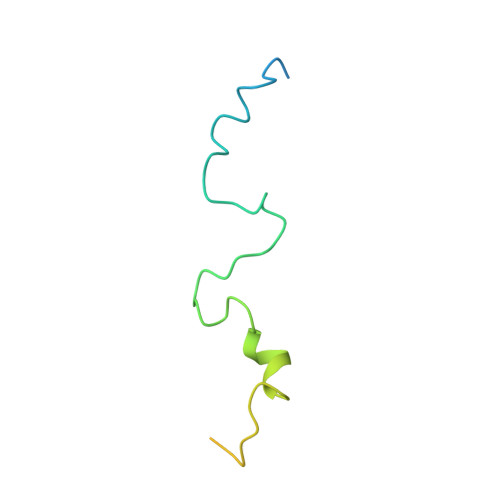
HMGB1-Facilitated p53 DNA Binding Occurs via HMG-Box/p53 Transactivation Domain Interaction, Regulated by the Acidic Tail.
Rowell, J.P., Simpson, K.L., Stott, K., Watson, M., Thomas, J.O.(2012) Structure 20: 2014-2024
- PubMed: 23063560
- DOI: https://doi.org/10.1016/j.str.2012.09.004
- Primary Citation of Related Structures:
2LY4 - PubMed Abstract:
Facilitated binding of p53 to DNA by high mobility group B1 (HMGB1) may involve interaction between the N-terminal region of p53 and the high mobility group (HMG) boxes, as well as HMG-induced bending of the DNA. Intramolecular shielding of the boxes by the HMGB1 acidic tail results in an unstable complex with p53 until the tail is truncated to half its length, at which point the A box, proposed to be the preferred binding site for p53(1-93), is exposed, leaving the B box to bind and bend DNA. The A box interacts with residues 38-61 (TAD2) of the p53 transactivation domain. Residues 19-26 (TAD1) bind weakly, but only in the context of p53(1-93) and not as a free TAD1 peptide. We have solved the structure of the A-box/p53(1-93) complex by nuclear magnetic resonance spectroscopy. The incipient amphipathic helix in TAD2 recognizes the concave DNA-binding face of the A box and may be acting as a single-stranded DNA mimic.
Organizational Affiliation:
Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK.