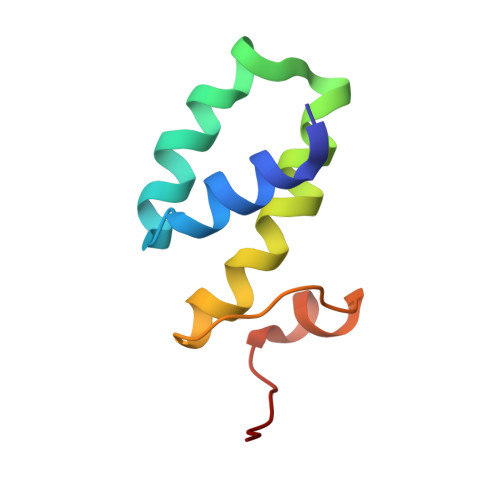
The high-precision solution structure of Yersinia modulating protein YmoA provides insight into interaction with H-NS
McFeeters, R.L., Altieri, A.S., Cherry, S., Tropea, J.E., Waugh, D.S., Byrd, R.(2007) Biochemistry 46: 13975-13982
- PubMed: 18001134
- DOI: https://doi.org/10.1021/bi701210j
- Primary Citation of Related Structures:
2K5S - PubMed Abstract:
The high-resolution solution structure of Yersinia modulating protein YmoA is presented. The protein is all helical with the first three of four helices forming the central core. Structures calculated with only NOE and dihedral restraints exhibit a backbone root-mean-square deviation (rmsd) of 0.77 A. Upon refinement against Halpha-Calpha, HN-N, and Calpha-C' J-modulated residual dipolar couplings, the backbone rmsd improves to 0.22 A. YmoA has a high amino acid sequence identity to and a similar overall fold to Escherichia coli hemolysin expression modulating protein Hha; however, structural differences do occur. YmoA is also found to be structurally similar to the histone-like nucleoid structuring protein H-NS, indicating that YmoA may intercalate into higher-order H-NS suprastructuring by substituting for an H-NS dimer.
Organizational Affiliation:
Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, USA.