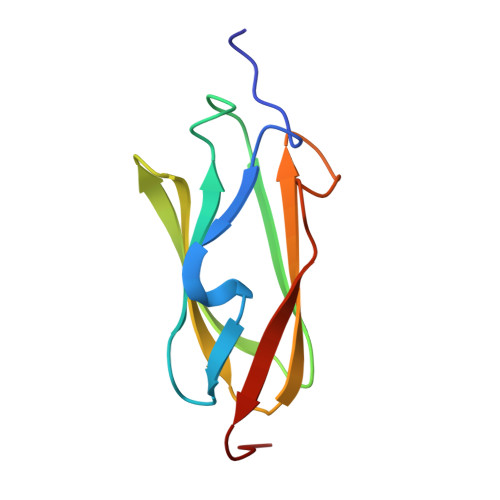
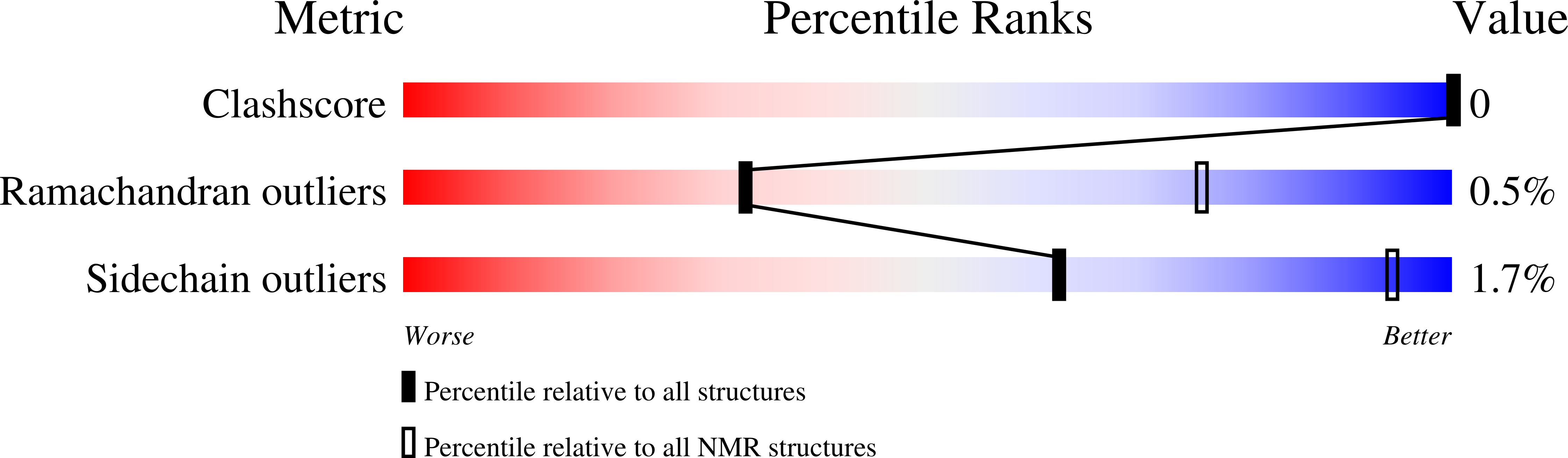Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations
Nakamura, F., Heikkinen, O., Pentikainen, O.T., Osborn, T.M., Kasza, K.E., Weitz, D.A., Kupiainen, O., Permi, P., Kilpelainen, I., Ylanne, J., Hartwig, J.H., Stossel, T.P.(2009) PLoS One 4: e4928-e4928
- PubMed: 19293932
- DOI: https://doi.org/10.1371/journal.pone.0004928
- Primary Citation of Related Structures:
2K3T - PubMed Abstract:
Mutations in filamin A (FLNa), an essential cytoskeletal protein with multiple binding partners, cause developmental anomalies in humans. We determined the structure of the 23rd Ig repeat of FLNa (IgFLNa23) that interacts with FilGAP, a Rac-specific GTPase-activating protein and regulator of cell polarity and movement, and the effect of the three disease-related mutations on this interaction. A combination of NMR structural analysis and in silico modeling revealed the structural interface details between the C and D beta-strands of the IgFLNa23 and the C-terminal 32 residues of FilGAP. Mutagenesis of the predicted key interface residues confirmed the binding constraints between the two proteins. Specific loss-of-function FLNa constructs were generated and used to analyze the importance of the FLNa-FilGAP interaction in vivo. Point mutagenesis revealed that disruption of the FLNa-FilGAP interface perturbs cell spreading. FilGAP does not bind FLNa homologs FLNb or FLNc establishing the importance of this interaction to the human FLNa mutations. Tight complex formation requires dimerization of both partners and the correct alignment of the binding surfaces, which is promoted by a flexible hinge domain between repeats 23 and 24 of FLNa. FLNa mutations associated with human developmental anomalies disrupt the binding interaction and weaken the elasticity of FLNa/F-actin network under high mechanical stress. Mutational analysis informed by structure can generate reagents for probing specific cellular interactions of FLNa. Disease-related FLNa mutations have demonstrable effects on FLNa function.
Organizational Affiliation:
Translational Medicine Division, Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA. fnakamura@rics.bwh.harvard.edu