Recognition of the H3K4me3 mark by the TAF3-PHD finger
van Ingen, H., van Schaik, F.M.A., Wienk, H., Ballering, J., Deschesne, A., Kruijzer, J., Timmers, H., Boelens, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription initiation factor TFIID subunit 3 | 75 | Mus musculus | Mutation(s): 0 Gene Names: Taf3 | 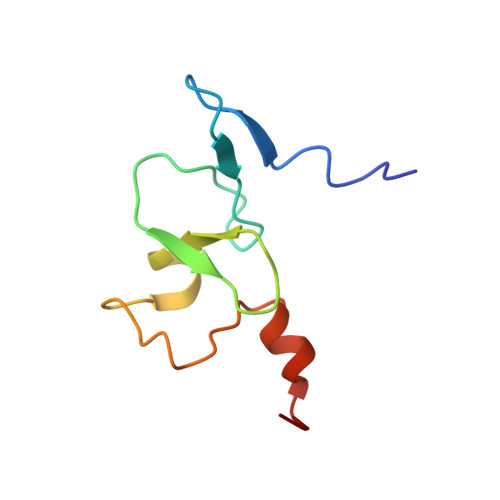 | |
UniProt | |||||
Find proteins for Q5HZG4 (Mus musculus) Explore Q5HZG4 Go to UniProtKB: Q5HZG4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5HZG4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | B [auth A], C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||