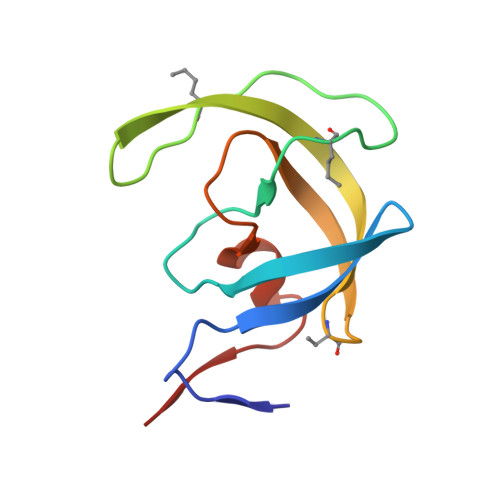
Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
Johnson, E.C.B., Malito, E., Shen, Y., Rich, D., Tang, W.J., Kent, S.B.(2007) J Am Chem Soc 129: 11480
- PubMed: 17705484
- DOI: https://doi.org/10.1021/ja072870n
- Primary Citation of Related Structures:
2JE4 - PubMed Abstract:
As part of our ongoing studies of the human immunodeficiency virus type 1 (HIV-1) protease enzyme, we set out to develop a modular chemical synthesis of the protein from multiple peptide segments. Our initial attempts were frustrated by the insolubility of intermediate peptide products. To overcome this problem, we designed a synthetic strategy combining the solubility-enhancing properties of C-terminal (Arg)n tags and the biological phenomenon of autoprocessing of the Gag-Pol polyprotein that occurs during maturation of the HIV-1 virus in vivo. Synthesis of a 119-residue peptide chain containing 10 residues of the reverse transcriptase (RT) open reading frame plus an (Arg)(10) tag at the C-terminus was straightforward by native chemical ligation followed by conversion of the Cys residues to Ala by Raney nickel desulfurization. The product polypeptide itself completed the final synthetic step by removing the C-terminal modification under folding conditions, to give the mature 99-residue polypeptide. High-purity homodimeric HIV-1 protease protein was obtained in excellent yield and had full enzymatic activity; the structure of the synthetic enzyme was confirmed by X-ray crystallography to a resolution of 1.07 A. This efficient modular synthesis by a biomimetic autoprocessing strategy will enable the facile synthesis of unique chemical analogues of the HIV-1 protease to further elucidate the molecular basis of enzyme catalysis.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Institute for Biophysical Dynamics, Ben-May Department for Cancer Research, The University of Chicago, Chicago, Illinois 60637, USA.