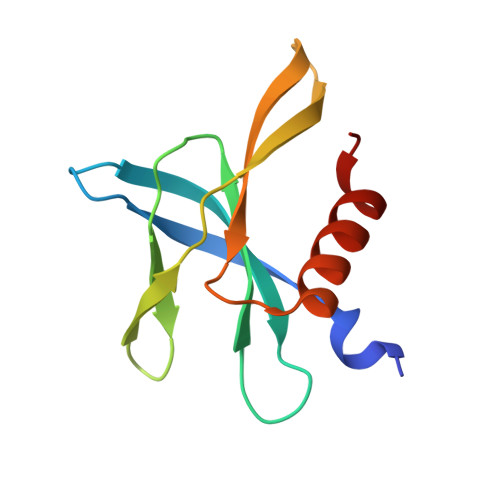
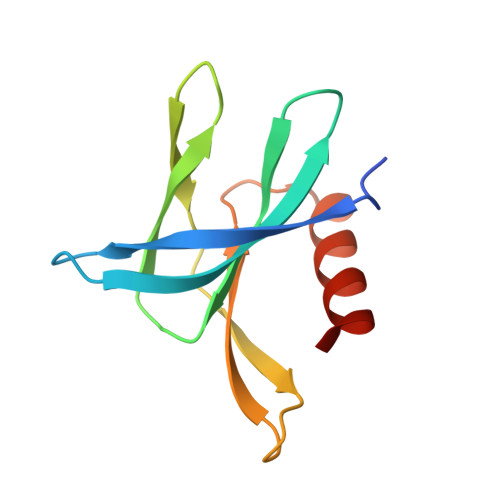
Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
Ponnusamy, R., Moll, R., Weimar, T., Mesters, J.R., Hilgenfeld, R.(2008) J Mol Biol 383: 1081
- PubMed: 18694760
- DOI: https://doi.org/10.1016/j.jmb.2008.07.071
- Primary Citation of Related Structures:
2J97, 2J98 - PubMed Abstract:
Non-structural protein 9 (Nsp9) of coronaviruses is believed to bind single-stranded RNA in the viral replication complex. The crystal structure of Nsp9 of human coronavirus (HCoV) 229E reveals a novel disulfide-linked homodimer, which is very different from the previously reported Nsp9 dimer of SARS coronavirus. In contrast, the structure of the Cys69Ala mutant of HCoV-229E Nsp9 shows the same dimer organization as the SARS-CoV protein. In the crystal, the wild-type HCoV-229E protein forms a trimer of dimers, whereas the mutant and SARS-CoV Nsp9 are organized in rod-like polymers. Chemical cross-linking suggests similar modes of aggregation in solution. In zone-interference gel electrophoresis assays and surface plasmon resonance experiments, the HCoV-229E wild-type protein is found to bind oligonucleotides with relatively high affinity, whereas binding by the Cys69Ala and Cys69Ser mutants is observed only for the longest oligonucleotides. The corresponding mutations in SARS-CoV Nsp9 do not hamper nucleic acid binding. From the crystal structures, a model for single-stranded RNA binding by Nsp9 is deduced. We propose that both forms of the Nsp9 dimer are biologically relevant; the occurrence of the disulfide-bonded form may be correlated with oxidative stress induced in the host cell by the viral infection.
Organizational Affiliation:
Institute of Biochemistry, Center for Structural and Cell Biology in Medicine, University of Lübeck, Ratzeburger Allee 160, 23538 Lübeck, Germany.