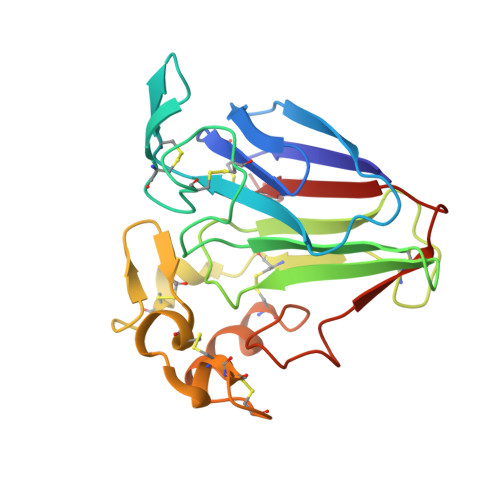
Crystal structure analysis of NP24-I: a thaumatin-like protein
Ghosh, R., Chakrabarti, C.(2008) Planta 228: 883-890
- PubMed: 18651170
- DOI: https://doi.org/10.1007/s00425-008-0790-5
- Primary Citation of Related Structures:
2I0W - PubMed Abstract:
The crystal structure of NP24-I, an isoform of the thaumatin-like protein (TLP) NP24 from tomato, has been reported. A prominent acidic cleft is observed between domains I and II of the three-domain structure of this antifungal protein, a feature common to other antifungal TLPs. The defensive role of the TLPs has also been attributed to their beta-1,3-glucanase activity and here too the acidic cleft is reported to play a vital role. NP24 is known to bind beta-glucans and so a linear beta-1,3-glucan molecule has been docked in the interdomain cleft of NP24-I. From the docked complex it is observed that the beta-glucan chain is so positioned in the cleft that a Glu and Asp residue on either side of it may form a catalytic pair to cause the cleavage of a glycosidic bond. NP24 has been reported to be an allergenic protein and an allergenic motif could be identified on the surface of the helical domain II of NP24-I. In addition, some allergenic motifs bearing high similarity/identity with some predicted Ig-E binding motifs of closely related allergenic TLPs like Jun a 3 (Juniperus ashei, from mountain cedar pollen) and banana-TLP have been identified on the molecular surface of NP24-I.
Organizational Affiliation:
Crystallography and Molecular Biology Division, Saha Institute of Nuclear Physics, 1/AF Bidhannagar, Kolkata 700 064, India.